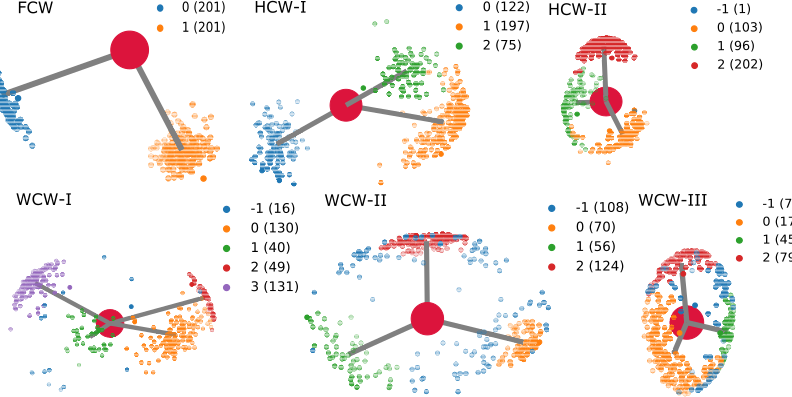
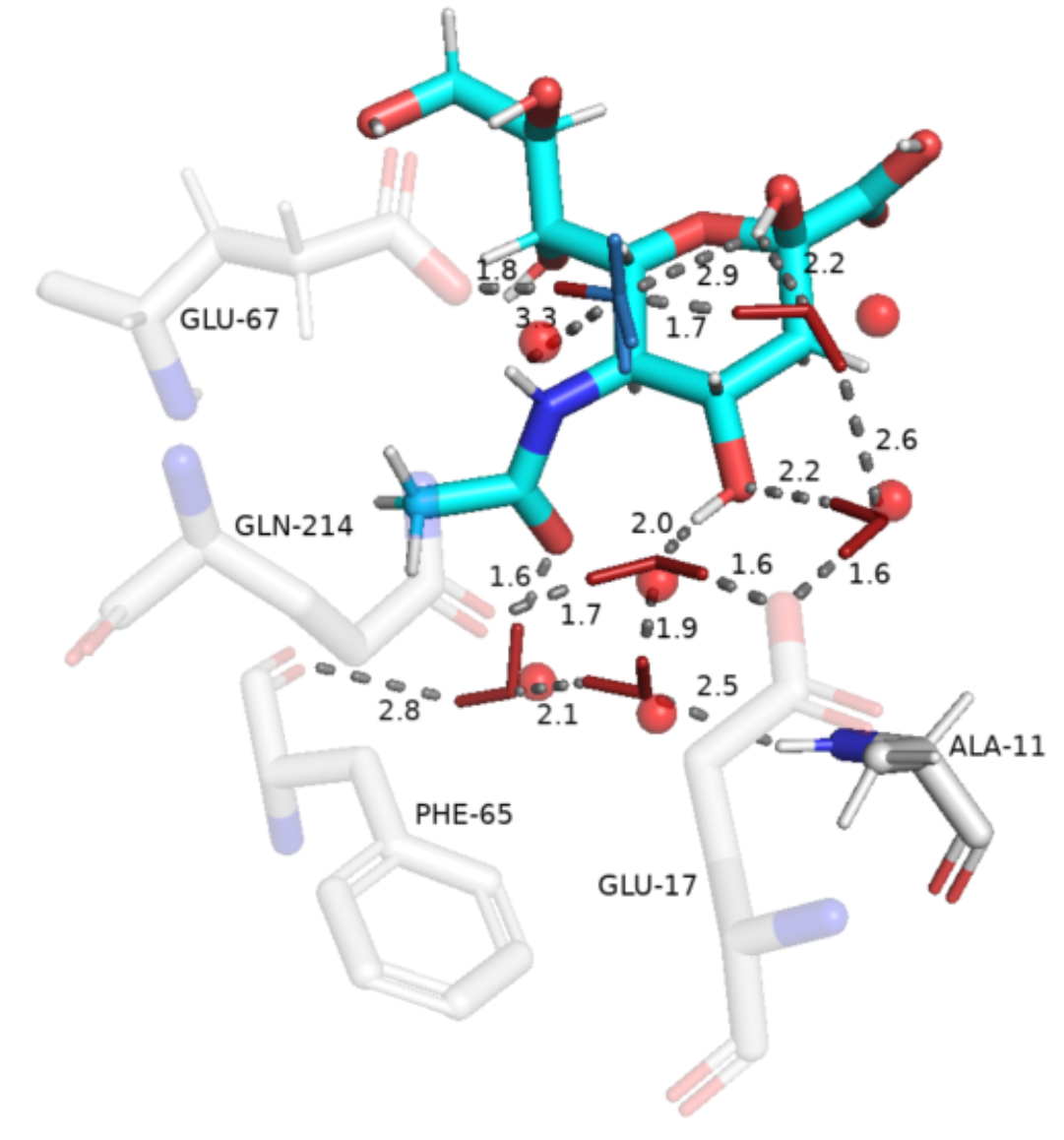
The WaterNetworkAnalysis (WNA) Python package serves as a set of tools for input preparation for ConservedWaterSearch python package from MD trajectories and their analysis. Using the ConservedWaterSearch package the conserved water molecules can be further classified into 3 distinct conserved water types based on their hydrogen orientation: Fully Conserved Waters (FCW), Half Conserved Waters (HCW) and Weakly conserved waters (WCW) - see figure below for examples. WNA can be used to create PyMol or nglview visualisations of conserved water networks for drug discovery or materials science purposes.
- Documentation: hosted on Read The Docs
- GitHub repository: source code/contribute code
- Issue tracker: Report issues/ request features
- ConservedWaterSearch: prepare trajectories and analyse results for/from conservedwatersearch
Coming soon.
The easiest ways to install WaterNetworkAnalysis is using pip:
pip install WaterNetworkAnalysisConda builds will be available soon.
The following example shows how to use WaterNetworkAnalysis to prepare a MD trajectory and analyse the results for determination of conserved water networks.
from WaterNetworkAnalysis import WaterNetworkAnalysis as WNA
import os
# MD trajectory filename
trajectory="md_pl.xtc"
# topology filename
topology="md_pl.gro"
# ligand name
ligand = 'SLB'
# distance to select water molecules around
distance = 15.0
# define active site by aminoacid residue numbers
active_site_aminoacids = [10,11,124,127,147,149,150,151,153,154,168,169,17,170,173,187,188,191,197,212,214,49,65,66,67,69,70,72]
analysis=WNA(aminoacids_in_activesite=active_site_aminoacids)
# if trajectory is not aligned align it and extract water molecules inside 15 A around active site
if not os.isfile('aligned_trajectory.xtc'):
analysis.align_trajectory(trajectory, topology,every=10)
analysis.extract_waters_from_trajectory(topologyology=topology, dist=distance)
elif not os.isfile('water_coordinates.dat'):
analysis.extract_waters_from_trajectory(traj='aligned_trajectory.xtc',topologyology=topology, dist=distance)
else:
analysis.load_H2O(fname='water_coordinates.dat')
# If the procedure hasn't started start it, else restart it or if finished load results
if not os.isfile('Clustering_results.dat'):
if not os.isfile('Clustering_results_temp.dat'):
analysis.cluster()
else:
analysis.restart_cluster()
else:
analysis.read_results()
# Make results in pdb file
analysis.make_results_pdb("aligned.pdb",ligand,mode="cathegorise")
analysis.make_results_pdb("aligned.pdb",ligand)
# create a PyMol visualisation session
analysis.visualise_pymol()