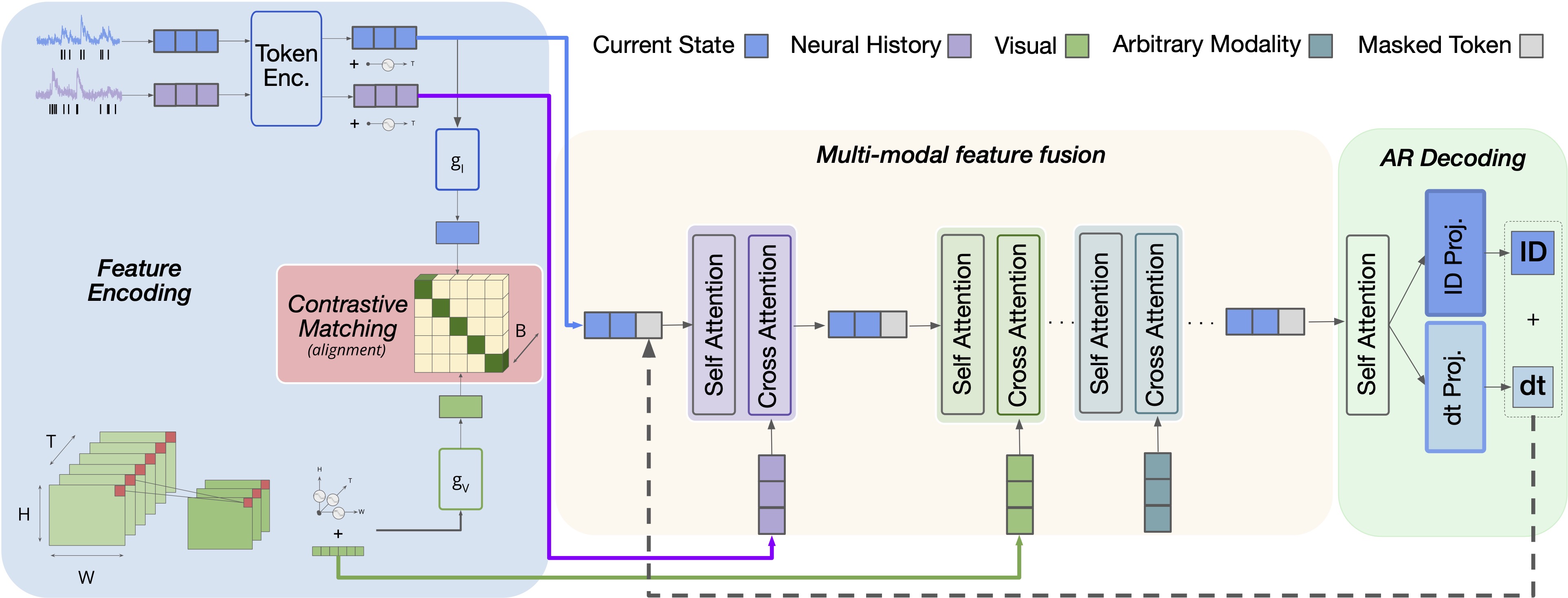
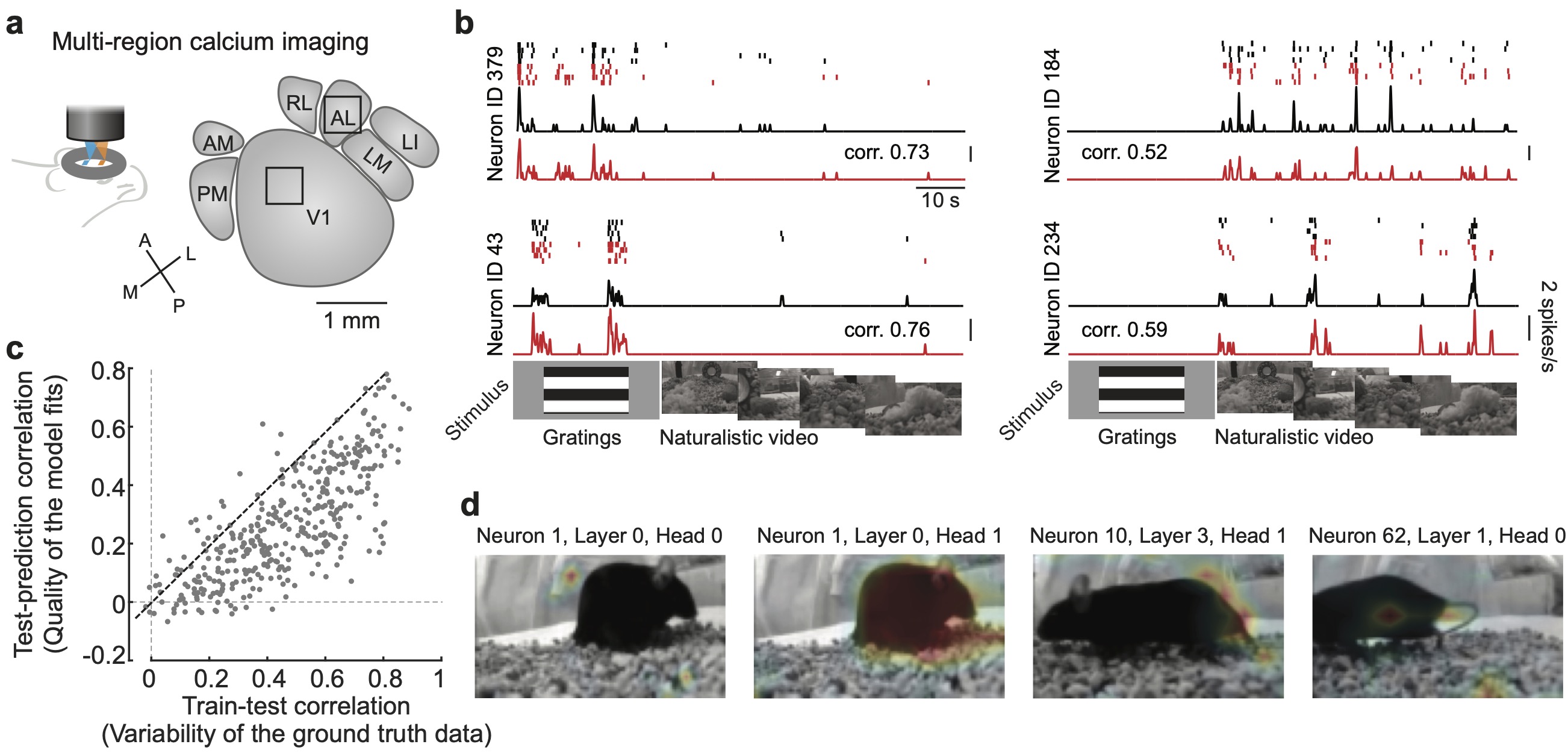
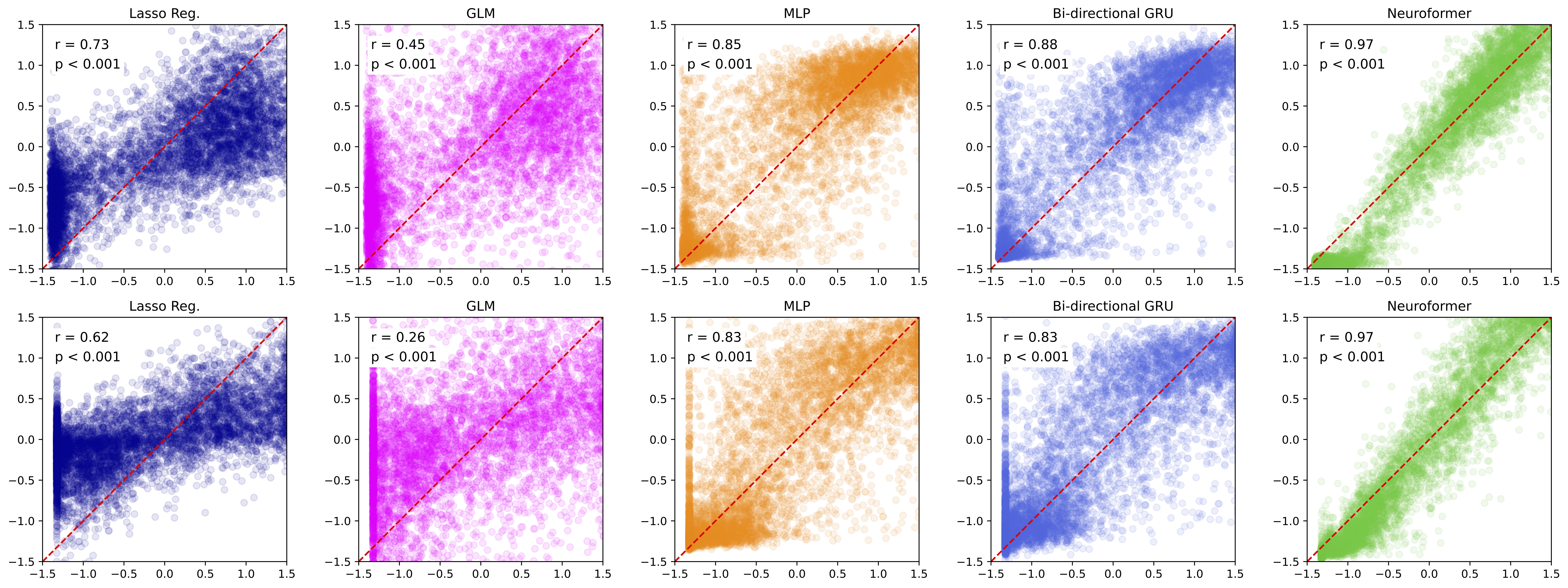
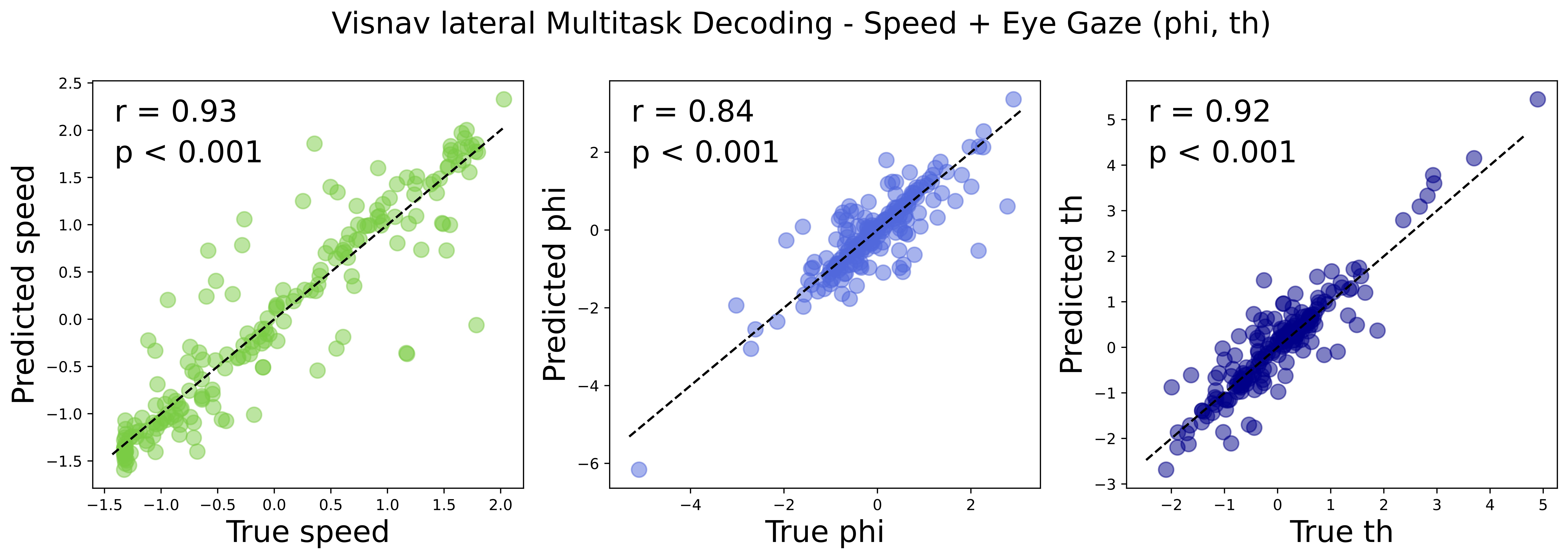
A GPT based multi-modal, multi-task transformer model for pretraining and downstream inference involving brain data.
Need help using the model with your data, or have general questions? Feel free to contact antonis@ucsb.edu.
conda create -n neuroformer
conda activate neuroformer
pip install -r requirements.txt
The Smith Lab has open-sourced two datasets for use with this model. Special thanks to Yiyi Yu and Joe Canzano 🙂
-
V1AL: Neuronal activity from the primary visual cortex and a higher visual area (V1 + AL), recorded from awake mice viewing visual stimuli. -
lateral(visnav): Recordings from the lateral visual cortex, spanning V1 and multiple higher visual areas, from mice engaged in a visually-guided navigation task. This dataset includes additional behavioral variables such as speed, and eye gaze (phi, th).
To pretrain on the visnav dataset, you can run the following code:
python neuroformer_train.py --lateral --config configs/NF/pretrain_visnav.yaml
For a closer look at the data format, refer to the [neuroformer.datasets.load_visnav()] function (used for example in the neuroformer_train.py).
{
'data': {
'spikes': (N_neurons, N_timesteps), # np.ndarray, required key
'frames': (N_frames, N_timesteps), # np.ndarray, optional key
'behavior variables': (N_timepoints,), # np.ndarray,
'intervals': (N_timepoints,), # np.ndarray, Denoting all intervals/time bins of the data. Used to split the data into train/val/test sets.
'train_intervals': (N_timepoints,) , # np.ndarray, The corresponding train intervals.
'test_intervals': (N_timepoints,) , # np.ndarray, The corresponding test intervals.
'finetune_intervals': (N_timepoints,) , # np.ndarray, The corresponding finetune intervals (very small amount).
'callback': callback() # function
}data['spikes']: Represents neuronal spiking data with dimensions corresponding to the number of neurons and timesteps.
data['frames']: If provided, it denotes stimulus frames that align with the neuronal spiking data.
data['behavior variables']: Optional key that represents behavioral variables of interest. The naming for this key can be customized as required.
data['behavior']: If provided, denotes the behavioral variable of interest (e.g. speed, phi, thi, etc). You can name this key as per your requirements and specify its usage in the config file (see below).
intervals: Provides a breakdown of time intervals or bins in the dataset.
train_intervals, test_intervals, finetune_intervals: Represent the segments of the dataset that will be used for training, testing, and fine-tuning respectively.
callback: This function is passed to the dataloader and parses your stimulus (for example how to index the video frames) according to the relationship it has to your response (spikes). It is designed to integrate any stimulus/response experiment structure. Typically requires only 4-5 lines of code; refer to comments inside visnav_callback() and combo3_V1AL_callback inside datasets.py for an example.
In the mconf.yaml file, you can specify additional modalities other than spikes and frames. For example behavioral features. The model will automagically create add/remove the necessary layers to the model. Additionally, you can specify any downstream objective, and choose between a
Here's what each field represents:
Modalities: Any additional modalities other than spikes and frames.
Modality Type: The name of the modality type. (for example behavior)
Variables: The name of the modality.
Data: The data of the modality in shape (n_samples, n_features).
dt: The time resolution of the modality, used to index n_samples.
Predict: Whether to predict this modality or not. If you set predict to false, then it will not be used as an input in the model, but rather to be predicted as an output.
Objective: Choose between regression or classification. If classification is chosen, the data will be split into classes according to dt.You can jointly pretrain the model using the spike causal masking (SCLM) objective and any other downstream task. The trainer will automatically save the model that does best for each corresponding objective (if you also include a holdout dataset). For example model.pt (normal pretraining objective), model_speed.pt, etc.
To finetune the model on one of the behavioral variables (speed, phi, thi), you can run the following code:
python neuroformer_train.py --lateral --finetune --loss_brop speed phi th --config configs/NF/finetune_visnav_all.yaml
--loss_bprop tells the optimizer which losses to backpropagate.
--config Here only difference between the two is adding Modalities.Behavior.Variables.(Data, dt, Predict, Objective) to the config file.
To generate new spikes:
python neuroformer_inference.py --dataset lateral --ckpt_path "model_directory" --predict_modes speed phi th
The neuroformer.utils.predict_modality() function can be used to generate predictions for any of the behavioral variables. See neuroformer_inference.py for an example.
# block_type = behavior, modality = speed
preds = predict_modality(model, dataset,
modality='speed',
block_type='behavior',
objective=config.modalities.behavior.speed.objective)
# 'regression' or 'classification'Note that if you want to generate predictions for a variable that was not used in the pretraining, you will need to add it to the config file (and preferably finetune on it first).