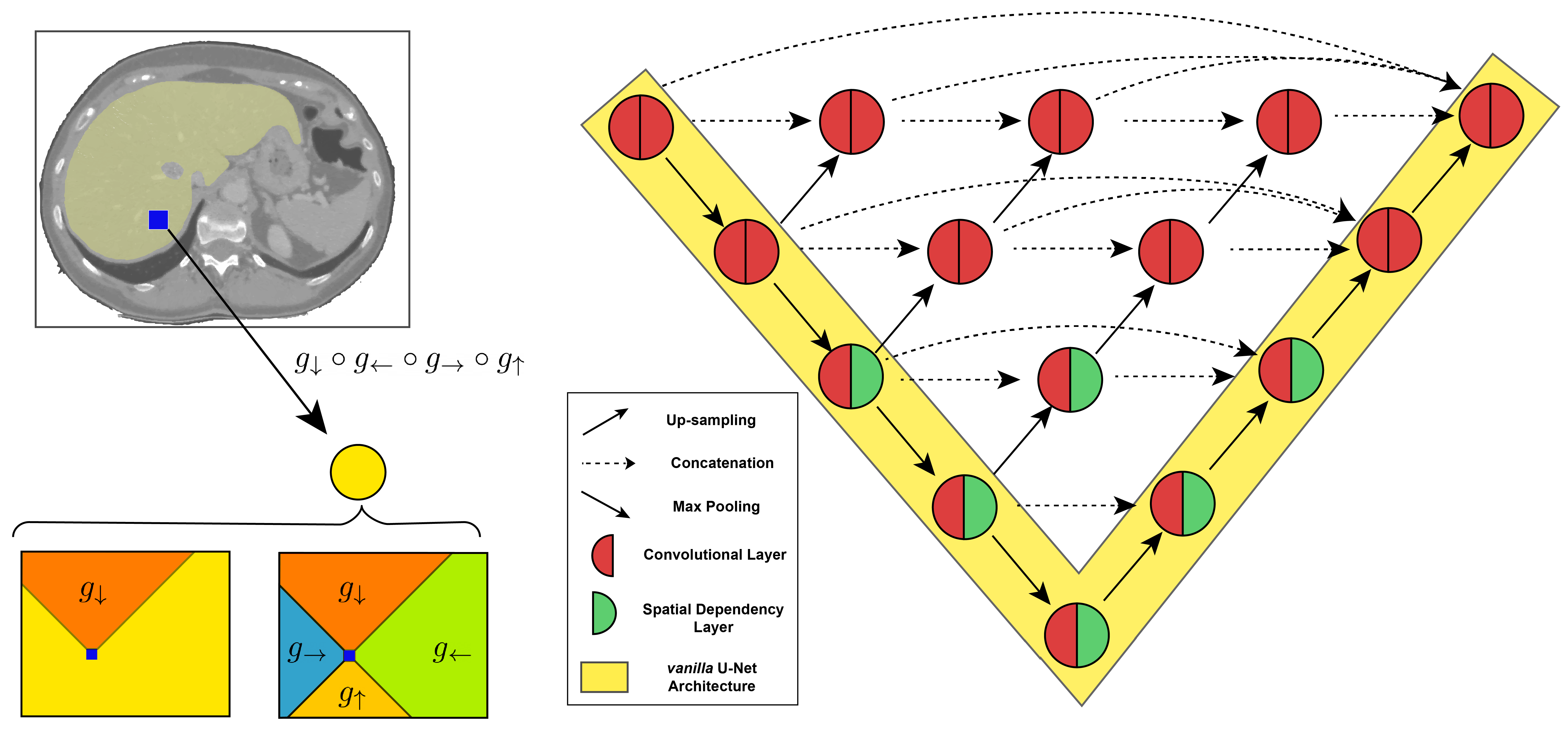
For medical image segmentation to generalize, we need two components: to identify local descriptions, but at the same time to develop a holistic representation of the image that captures long-range spatial dependencies. Unfortunately, we demonstrate that the start of the art does not achieve the latter.
We introduce a novel deep neural network architecture endowed with spatial recurrence that achieves this. The implementation relies on gated recurrent units, and in particular on spatial dependency layers, that directionally traverse the feature map, greatly increasing each layers receptive field and explicitly modeling non-adjacent relationships between pixel.
This implementation is competitive with state of the art methods, surpassing multiple transformer based architectures.
You can find the paper that accompanies this work here: Holistic Modeling in Medical Image Segmentation Using Spatial Recurrence
.
├── data/
├── dsb2018/ # where DSB2018 data is stored
├── lits/ # where LiTS data is stored
├── polips/ # where CVC_ClinicDB data is stored
├── segTHOR.py # where SegTHOR data is stored
├── figs/ # figures from the paper
├── lib/
├── core.py # building blocks and parent LightningModule for all models
├── dataset.py # Pytorch/PytorchLightning specific modules for the datasets anda dataloaders
├── models.py # all architectures
├── sdn.py. # SDN modules
├── utils.py # utility with helper functions, loss functions and evaluation metrics
├── train.py # generic training script
├── requirements.txt
├── LICENSE
└── README.md
To reproduce the experiments described in the paper you will first need to install PyTorch and PyTorchLightning. To fully reproduce the experiments on the segTHOR dataset you will need a system equipped with a TeslaV100 GPU with 32Gb of memory, otherwise, reduce the batch size when training at a cost of segmentation performance.
To install all the requirements use:
pip install -r requirements.txtTo try specific experiments run:
python train.py --dataset DATASET_NAME --train True --test True --n_gpus 1 --early_stopping True --batch_size N --lr 0.0001 --block vgg --model MODEL_NAME --n_layers M --exp_name EXP_NAMEReplacing DATASET_NAME, MODEL_NAME, EXPERIMENT_NAME, N, and M approprietly for the configurations of the experiment.
data downloaded at: https://www.kaggle.com/c/data-science-bowl-2018/data
[Haghighi, M., Heng, C., Becker, T., Doan, M., McQuin, C., et al.: Nucleus Segmentation Across Imaging Experiments: The 2018 Data Science Bowl. Nature Methods 16(12), 1247–1253 (2019]
database description and download link available at: https://polyp.grand-challenge.org/CVCClinicDB/
[Bernal, J., Tajkbaksh, N., Sánchez, F.J., Matuszewski, B.J., Chen, H., Yu, L.,Angermann, Q., Romain, O., Rustad, B., Balasingham, I., et al.: Comparative Validation of Polyp Detection Methods in Video Colonoscopy: Results From the Miccai 2015 Endoscopic Vision Challenge. IEEE Transactions on Medical Imaging 36(6), 1231–1249 (2017)]
database description and download instructions at: https://competitions.codalab.org/competitions/17094
[Bilic, P., Christ, P.F., Vorontsov, E., Chlebus, G., Chen, H., Dou, Q., Fu, C., Han,X., Heng, P., Hesser, J., Kadoury, S., Konopczynski, T.K., Le, M., Li, C., Li, X.,Lipková, J., Lowengrub, J.S., Meine, H., Moltz, J.H., Pal, C., Piraud, M., Qi, X.,Qi, J., Rempfler, M., Roth, K., Schenk, A., Sekuboyina, A., Zhou, P., Hülsemeyer,C., Beetz, M., Ettlinger, F., Grün, F., Kaissis, G., Lohöfer, F., Braren, R., Holch,J., Hofmann, F., Sommer, W.H., Heinemann, V., Jacobs, C., Mamani, G.E.H.,van Ginneken, B., Chartrand, G., Tang, A., Drozdzal, M., Ben-Cohen, A., Klang,E., Amitai, M.M., Konen, E., Greenspan, H., Moreau, J., Hostettler, A., Soler, L.,Vivanti, R., Szeskin, A., Lev-Cohain, N., Sosna, J., Joskowicz, L., Menze, B.H.: The Liver Tumor Segmentation Benchmark (LiTS). CoRRabs/1901.04056(2019)]
database description and download link available at: CodaLab - Competition
[Zoé Kanvertm Caroline Petitjean, Bernard Dubray, Su Ruan]