PatchAlign : Fair and Accurate Skin Disease Image Classification by Alignment with Clinical Labels [MICCAI 2024]
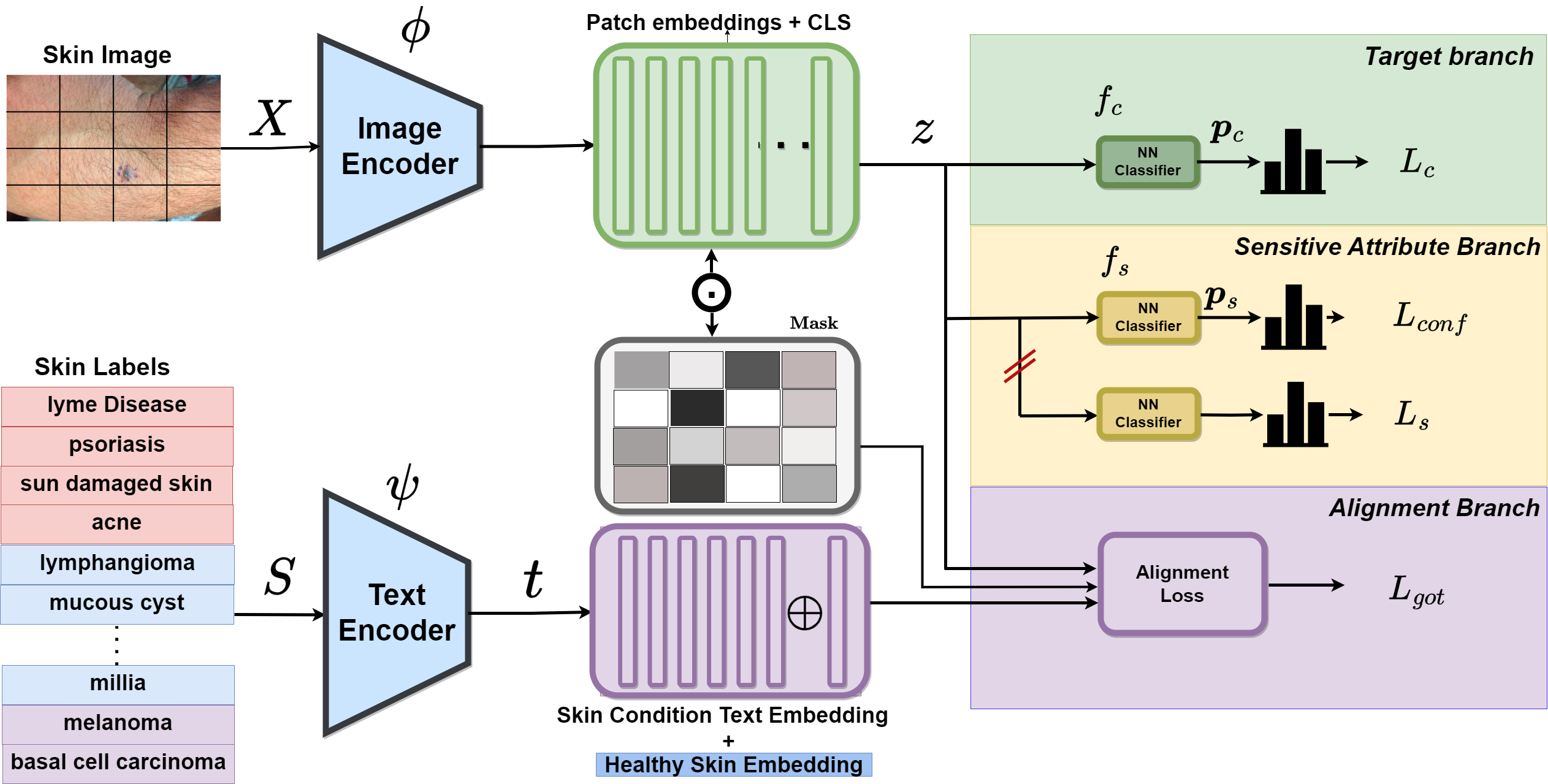
PatchAlign: Our proposed alignment-based skin disease classifier
The libraries used and their version requirements for running the code are given in requirements.txt. Please create a virtual environment (conda/pyvenv) to avoid any version conflicts.
pip install -r requirements.txt-
Download Fitzpatrick17k dataset by filling the form here
-
Download Diverse Dermatology Images (DDI) from here
-
Use data_play_fitz.ipynb and data_play_ddi.ipynb to remove unknown skin types, encode disease labels, and generate the weights of reweighting and resampling methods.
-
Change the fitzpatrick skin-tones number in ddi_metadata_code.csv from 1,2,3 to 1,3,5 respectively before running the code.
-
Note: For Text Embeddings use 'text_embeddings_3_large_consecutive_averaged.npy' directly or use create_embeddings.ipynb to test other ways of creating text embeddings.
Run the following comands to reproduce the results. Before running the training code, run create_class_indices.ipynb.
# 0 - In Domain, 1 - Out Domain A12, 2 - Out Domain A34, 3 - Out Domain A56
# PatchAlign DDI In-Domain Result
python3 -u train_PatchAlign_DDI_InDomain.py 20 full ddi DDI_INDOMAIN
# PatchAlign FitzPatrick17k In-Domain
python3 -u train_PatchAlign_FitzPatrick_InDomain.py 20 full fitzpatrick PATCHALIGN_FITZ_INDOMAIN
# PatchAlign FitzPatrick17k Out-Domain Result (2 here means A56)
python3 -u train_PatchAlign_FitzPatrick_OutDomain.py 20 full fitzpatrick PATCHALIGN_FITZ_OUTDOMAIN 2
# For Other Results like Fair DisCo, eg: (here 0 means in Domain)
python -u train_DisCo.py 20 full fitzpatrick FairDisCo 0;
python -u train_BASE.py 20 full fitzpatrick Base 0;
# etc.For running the multitask files, first pass the metadata file through create_class_indices_multitask.ipynb ('ddi_metadata_code.csv' for ddi dataset and 'fitzpatrick17k_known_code.csv' for fitzpatrick dataset).
For DDI Dataset:
Command: python3 train_DisCo_multitask_ddi.py 25 full ddi multitask_ddi 32 1234
For Fitzpatrick Dataset:
Command: python3 train_DisCo_multitask_fitz.py 25 full fitzaptrick multitask_fitz 32 1234
Use multi_evaluate.ipynb
Please cite the paper and star this repository if you use our work. Queries regarding the paper or the code may be directed to aayushman20@iiserb.ac.in. Alternatively, you may also open an issue on this repository.
@misc{aayushman2024patchalignfairaccurateskindisease,
title={PatchAlign:Fair and Accurate Skin Disease Image Classification by Alignment with Clinical Labels},
author={Aayushman and Hemanth Gaddey and Vidhi Mittal and Manisha Chawla and Gagan Raj Gupta},
year={2024},
eprint={2409.04975},
archivePrefix={arXiv},
primaryClass={cs.CV},
url={https://arxiv.org/abs/2409.04975},
}- This code was built on top of the codebase of FairDisCo, GOT. We thank the developers of the Fitzpatrick17k dataset and DDI dataset for providing the dataset. FairDisCo for providing the baseline.