Annotation-layer for the INSDC BioSample database
The BioSample database contains descriptive meta-data for all biological samples housed by the "International Nucleotide Sequence Database Collaboration", the world's central repository for biological sequence data.
Due to the diversity of available samples and their descriptions, the meta-data is not standardized. Each record is stored as an XML file, containing its own set of tags and values.
BioAnnotate provides an annotation layer for BioSample to aggregate similar tags and standardize the value formats.
There are 42,125 unique 'tags' across >10.7 million BioSample XML files. We will annotate these tags into 4 categories, to allow for data-aggregation and ultimately a "clean" database.
geo: Geographic names and spatial coordinatesdate: Sample collection date and/or release dateorganism: Host or Pathogen speciesecosystem: Environmental origin description or body-site
We will work on a collaborative Annotation Spreadsheet which contains every unique BioSample tag.
Sign-up on the Lockout sheet to annotate a 'chunk' of 2,500 rows in the biosample_tags sheet for a particular class of data (see below).
The default for all tags is set to F for "FALSE". If a biosample_tag describes a field which is pertinent to your data-class, change this value to T for "TRUE".
If you are unsure of how to classify a particular biosample_tag, set the value to ? and/or ask in the chat.
Kat would like to annotate Chunk C for geo data.
-
She reviews the
geodata class description below to understand the inclusion and exclusion criteria for this data-class. -
She enters her name on the
Lockoutsheet to indicate she has begun to work on this chunk. -
On the
biosample_tagssheet Chunk corresponds to Rows5001 - 7500and thegeo_nameandgeo_coordcolumns. -
After turning on some good jams, she annotates these rows.
-
Upon completing her annotation, she updates
Lockoutto indicate this chunk is complete and she can begin working on another Chunk.
Inclusion: Tags which can provide any location data. Imagine the keywords you could type into Google Maps. e.g. geo_location, country, national_park, sequencing_institute, lake_name, longitude, lat_long, geo_coordinates...
Exclusion: Tags which describe a generic environment, not geographically specific. e.g. snow_depth, nitrogen_content_soil, lake_type...
-
geo_name: Set toTif tag likely contains words describing geo-data. -
geo_coord: Set toTif tag likely contains numbers describing geo-data, mainly longitude / latitude / altitude.
Inclusion: Tags which would contain a date. e.g. collection_date, sample_date, sequencing_date, release_date...
Exclusion: Tags which contain time-course data, such as the timeline of an experiment. e.g. week_of_growth, hours...
-
collection_date: Set toTif tag specifically describes the time at which a sample was collected from nature. -
other_date: Set toTif tag contains a date.
Inclusion: Tags which can provide taxonomic information regarding the organism which had been sampled. e.g. species, genus, scientific_order, taxonomy_string
Exclusion: Tags which describe a generic component of an organism. e.g. leaf_type, fur_colour, paw_length...
-
host_species: Default choice to set toTfor this class -
virus_species: Set toTif tag specifically indicates a viral organism classification.
Inclusion: Tags which can provide an environmental or organism-tissue description of the samples origin. e.g. water_depth, wastewater_site, soil_moisture, brain_region, tumour_diameter, organ_site...
-
ecosystem: Set toTif tag describe the samples environment. -
bodysite: Set toTif tag describes an organism's site.
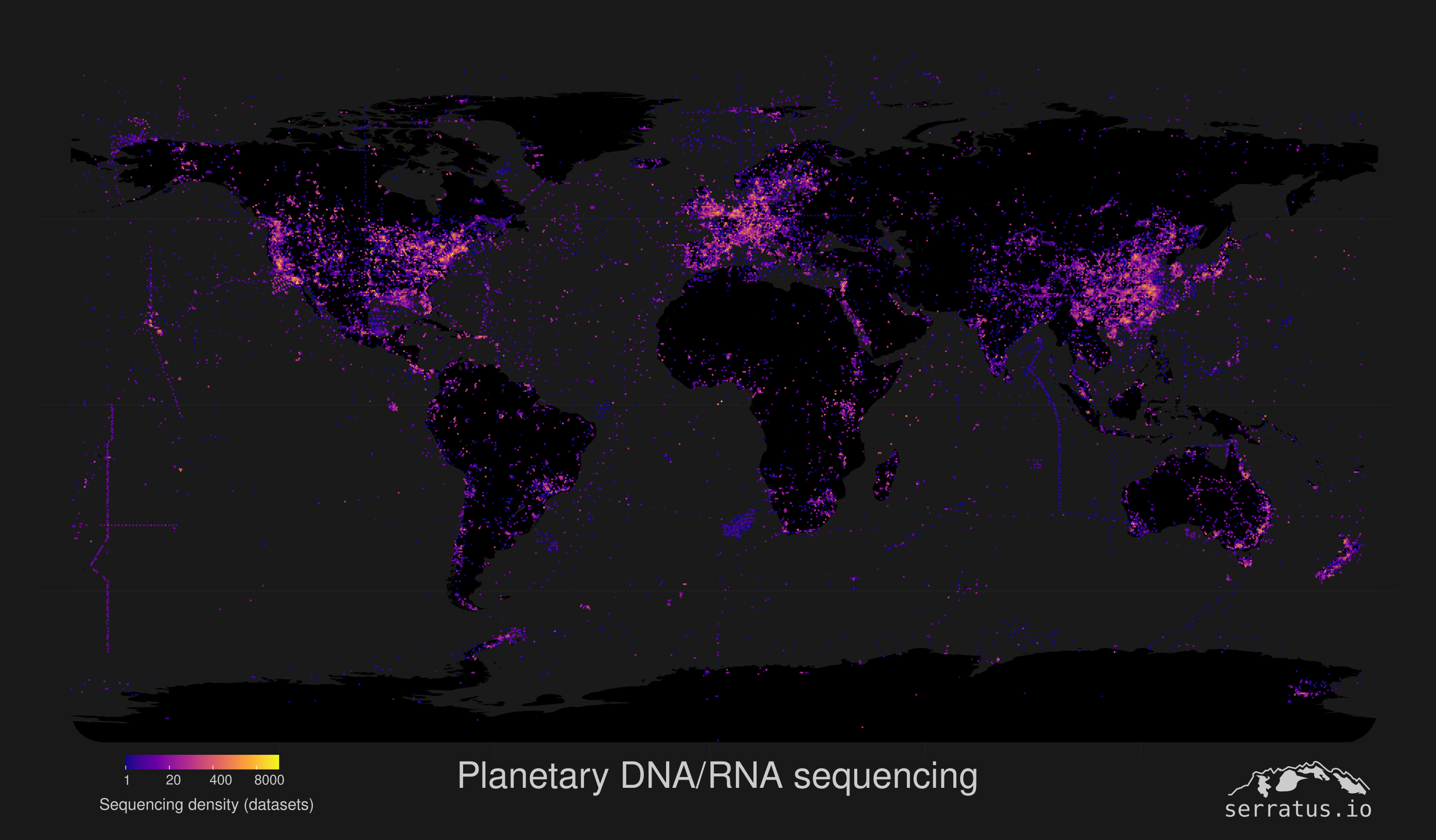
An example use-case for this data showing the spatial-distribution of several million DNA or RNA sequencing datasets in the 'Sequence Read Archive'. Geographic data was extracted from BioSample, but >50% of the data is missing due to inconsitent naming, we're going to fix that!
@ababaian @adrianbele @cbenon @linzzasaurus @mamurak @rgodinezp @schen1 @shiwanibiradar