genomic_seg_plots is a short pipeline to build genomic plots from genbank (short) genomes, displaying NCBI seg analysis output, and genomic features. It's main objective is to automatize and easily locate simple sequences (SS).
These are genomic_seg_plots dependencies, links to their installing instructions, commands for installation, and used versions:
| Software name (version used) | Terminal | Installation *debian based distros |
|---|---|---|
| tidyverse (1.3.0) | R (4.0.3) | install.packages("tidyverse") |
| ggrepel (0.9.1) | R (4.0.3) | install.packages("ggrepel") |
| NCBI seg | bash | download via ftp, compile and export permanently to $PATH |
It also assumes properly functioning perl (tested with v5.30.0), and seg working under any location. For instance, this is the expected output of entering seg at commnand line:
Usage: seg <file> <window> <locut> <hicut> <options>
<file> - filename containing fasta-formatted sequence(s)
<window> - OPTIONAL window size (default 12)
<locut> - OPTIONAL low (trigger) complexity (default 2.2)
<hicut> - OPTIONAL high (extension) complexity (default locut + 0.3)
<options>
-x each input sequence is represented by a single output
sequence with low-complexity regions replaced by
strings of 'x' characters
-c <chars> number of sequence characters/line (default 60)
-m <size> minimum length for a high-complexity segment
(default 0). Shorter segments are merged with adjacent
low-complexity segments
-l show only low-complexity segments (fasta format)
-h show only high-complexity segments (fasta format)
-a show all segments (fasta format)
-n do not add complexity information to the header line
-o show overlapping low-complexity segments (default merge)
-t <maxtrim> maximum trimming of raw segment (default 100)
-p prettyprint each segmented sequence (tree format)
-q prettyprint each segmented sequence (block format)
To clone this repo via command line git, enter the following commands:
git clone https://github.com/abelardoacm/genomic_seg_plots.git
cd genomic_seg_plots/bin/
chmod +x *
if all requirements are met you should be ready to go
.
├── bin <- Where you have to be to invoke scripts
│
├── data
│ └── Raw_database <- Place here genomic genbank files
│
└── results
├── GenFeatures_locations <- Contains csv files with genomic features extracted from genbank file
├── Proteomic_fasta <- Fasta aminoacid files to serve as seg input
├── seg <- NCBI seg analysis output for high (.faa), low (.faa) and both (.csv) type of sequences
├── Complexity_genomic_plots/anytaxon <- tiff figures built with ggplot2
├── stackedplots <- tiff files of stacked SS plots
Analyzed sequences of anytaxon must be contained in data/Raw_Database/ for what we suggest to download sequences taxon by taxon as follows, with the general query:
anytaxon [ORGANISM] AND srcdb_refseq[PROP]
From bin you only have to invoke one of included scripts,
./genomic_slimcomplots.sh <anytaxon> <windowsize> <complexity low cut> <complexity high cut>
for example (with included data):
./genomic_slimcomplots.sh Nidovirales 12 1.9 2.1
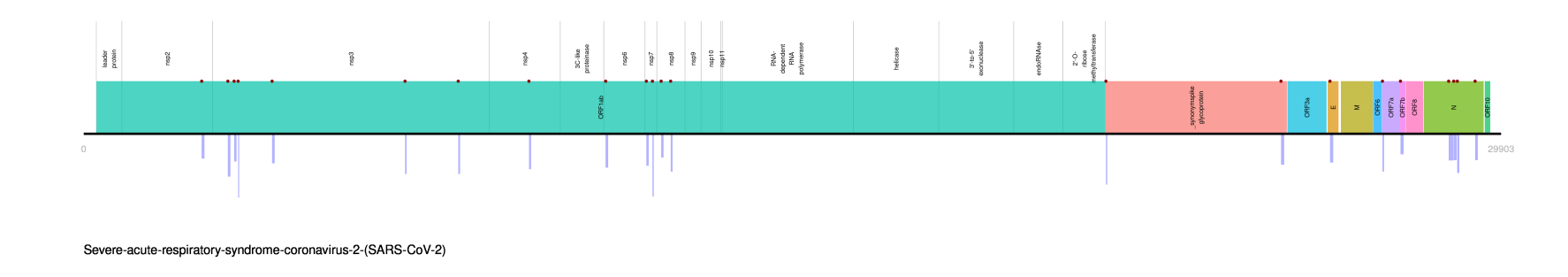
will save several genomic plots at results/Complexity_genomic_plots/ anytaxon subfolder, indicating simple sequences in genome.
This is the genomic slimcompplot for SARS CoV2: