Predictive models for tumor growth, and tools to apply them to clinical data
Please refer to the documentation for an overview of this package.
Code snippet:
using TumorGrowth, Lux, Random
times = [0.1, 6.0, 12.0, 18.0, 25.0, 30.0, 35.0, 41.0, 47.0]
volumes = [0.013, 0.0072, 0.0043, 0.0021, 0.0043, 0.0043, 0.0044, 0.0058, 0.015]
# define an experimental model based on a neural ODE:
network = Lux.Chain(Dense(2, 3, Lux.tanh; init_weight=Lux.zeros64), Dense(3, 2))
neural_model = neural2(Random.default_rng(), network)
# compare with with some classical models:
models = [neural_model, logistic, bertalanffy]
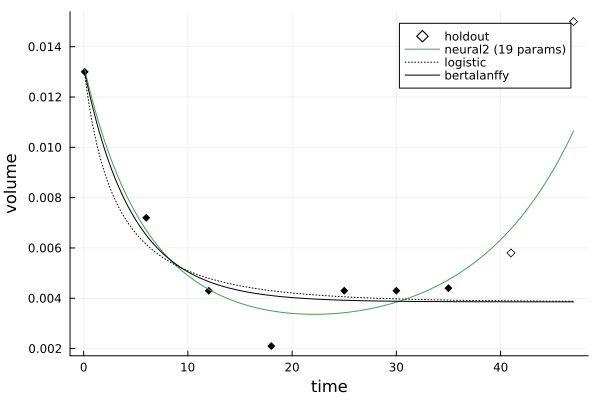
julia> comparison = compare(times, volumes, models, holdouts=2)
ModelComparison with 2 holdouts:
metric: mae
neural2 (19 params): 0.002656
logistic: 0.00651
bertalanffy: 0.006542
using Plots
julia> plot(comparison)The datasets provided the TumorGrowth.jl software are sourced from Laleh et al. (2022) "Classical mathematical models for prediction of response to chemotherapy and immunotherapy", PLOS Computational Biology", with some restructuring provided by Yasin Elmaci and Okon Samuel.