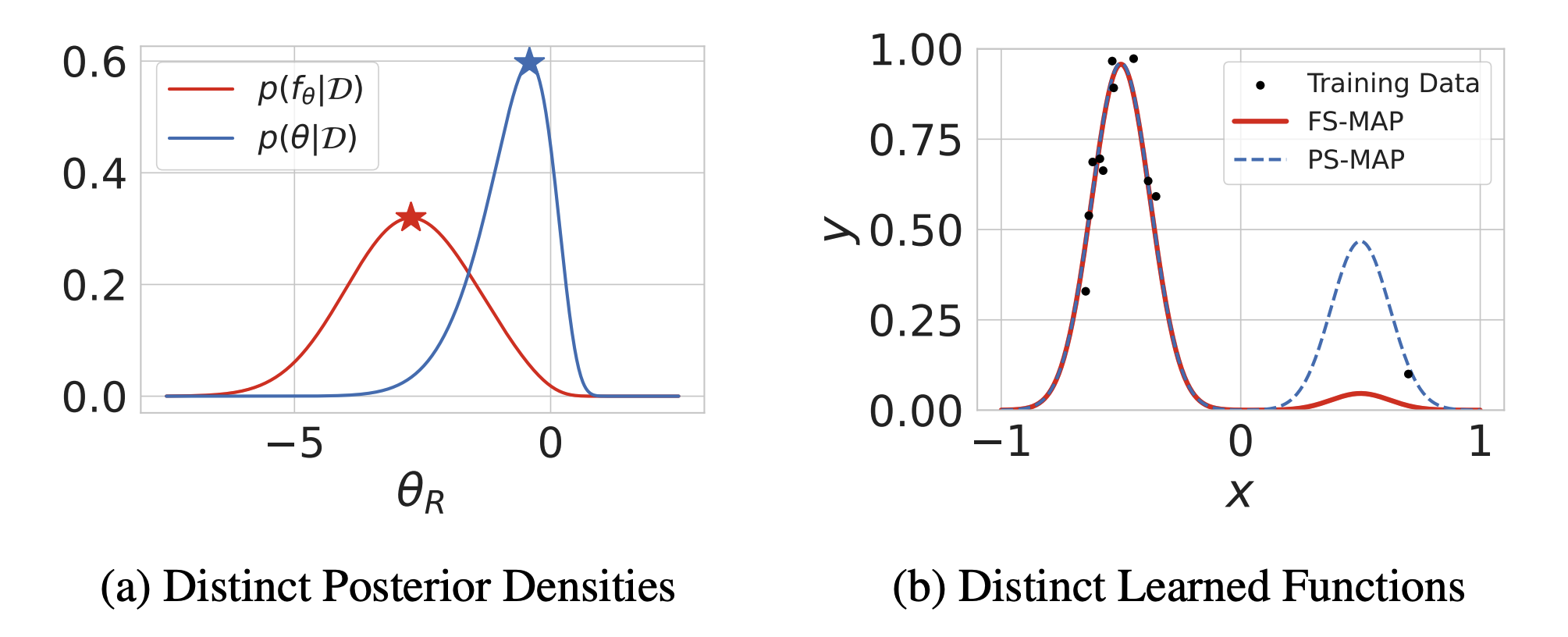
This repository contains the code for the paper Should We Learn Most Likely Functions or Parameters? by Shikai Qiu, Tim G. J. Rudner, Sanyam Kapoor and Andrew Gordon Wilson (NeurIPS 2023).
Standard regularized training procedures correspond to learning the most likely parameters of our model given the data and our prior. However, model parameters are of interest only insomuch as they combine with the functional form of a model to provide a function that can make good predictions. Moreover, the most likely parameters under the parameter posterior do not generally correspond to the most likely function induced by the parameter posterior. The aim of this paper is to improve our understanding of what it means to learn most likely functions instead of parameters. We investigate the benefits and drawbacks of directly estimating the most likely function implied by the model and the data. We show that this procedure leads to pathological solutions when using neural networks, and prove conditions under which the procedure is well-behaved, as well as a scalable approximation. Under these conditions, we find that function-space MAP estimation can lead to flatter minima, better generalization, and improved robustness to overfitting. On the other hand, while distinguishing between the two approaches proves valuable when we have highly motivated priors, the advantages are modest in many practical problems with uninformative priors.Create a new conda environment (if needed):
conda env create -f environment.yml -n fsmap
The codebase has been tested with the latest PyTorch.
pip install torch torchvision --extra-index-url https://download.pytorch.org/whl/cu118/Install CUDA-compiled JAX version from here. The
codebase has been tested with JAX version 0.4.
pip install "jax[cuda11_pip]" -f https://storage.googleapis.com/jax-releases/jax_cuda_releases.htmlNOTE: Use cuda11_pip or cuda12_pip depending on CUDA versions.
And finally, run
pip install -e .
-
Gaussian Mixture Model Regression (Sections 1 & 4.3)
Run./notebooks/gaussian_mixture.ipynb -
Fourier Basis Regression (Section 3.3)
Run./notebooks/fourier.ipynb -
Visualizing Lack of Reparameterization Invariance of PS-MAP (Appendix A)
Run./notebooks/reparam.ipynb
Run ./notebooks/uci.ipynb
The main file for training PS-MAP is experiments/train.py.
An example command to run the training with FashionMNIST with PS-MAP is
python experiments/train.py \
--dataset=fmnist --batch-size=128 \
--model-name=resnet18 \
--lr=0.1 --weight-decay=5e-4 \
--epochs=50 --seed=173 \
--log-dir=logsSee below for a summary of all important arguments:
--dataset: The training dataset. e.g.fmnist(FashionMNIST),cifar10(CIFAR-10), etc.--ctx-dataset: The context dataset.--model-name: We useresnet18(ResNet-18) for all our experiments.--batch-size: Size of the minibatch used for each gradient update.--context-size: Size of the minibatch used for each context gradient update.--epochs: Number of epochs to train for.--lr: Learning rate for SGD.--weight-decay: Weight decay for SGD.--reg_scale: Regularization scale for use withL-MAP. Default0.0corresponds toPS-MAP.--seed: Seed used for model initialization, and dataset sampling.
We use pretrained ResNet checkpoints from Flaxmodels.
To use them, simply specify --model-name=resnet50_pt or --model-name=resnet18_pt in conjunction with other arguments above.
Note that when using with CIFAR-10, we need to resize the images to 224 x 224 to be compatible with pretraining. We provide the resized versions via the argument --dataset=cifar10_224.
We specify context dataset for L-MAP as --ctx-dataset=cifar100_224.
The file experiments/evaluate.py can be used to evaluate trained checkpoints independently.
The key arguments are:
--model-name: We useresnet18(ResNet-18) for all our experiments, same as above.--ckpt-path: Path to the directory where the checkpoint is stored, or the checkpoint directly. This is often the prefix of the--log-dirreported in the training runs.--dataset: The dataset with which the checkpoint was trained (so that the checkpoint parameters can be mapped correctly).--batch-size: Batch size used during evaluation. A larger batch size can be used since we do not need memory to preserve computational graphs.
The file experiments/evaluate_landscape.py can be used to assess the landscape around trained checkpoints. We use Filter Normalization to plot perturbations along random directions around the optimum.
In terms of design, the number of GPUs avaiable decides the number of random directions we evaluation in a single run (via pmap),
and on each GPU we take multiple steps in the random directions evaluated (via vmap).
A sample command to evaluate CIFAR-10 landscape is:
python experiments/evaluate_landscape.py \
--dataset=cifar10 --model-name=resnet18 \
--batch-size=128 --ckpt-path=${CKPT_PATH} \
--step_lim=50. --n-steps=50 \
--seed=137The new key arguments are:
--step-lim: The limit of the step sizes, such that we take steps along the random direction as[-s, s].--n-steps: The number of steps in the range[-s,s].
Use CUDA_VISIBLE_DEVICES to control the number of GPUs available for parallel evaluation of multiple random directions.
Alternatively, use multiple runs to get more random directions with different seeds.
Apache 2.0
Please cite our work as:
@inproceedings{qiu2023should,
title={{Should We Learn Most Likely Functions or Parameters?}},
author={Shikai Qiu and Tim G. J. Rudner and Sanyam Kapoor and Andrew Gordon Wilson},
booktitle={Advances in Neural Information Processing Systems},
year={2023}
}