For use with MRI data
Release notes (version 2.5)
- Add a check for the nifti directory when the dicom directory does not contain any session folder
Ensures that the dicoms are unpacked to the correct session folder for subjects with multiple sessions
-
Convert dicoms to BIDS using dcm2niix and without the need of a
.jsonconfiguration file -
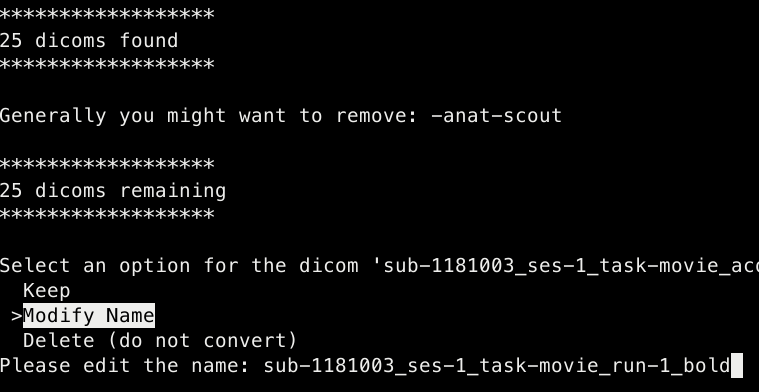
Script converts all dicoms and then leads the user through a renaming and moving files process
-
Script adds the intended for field to the fieldmap
.json -
The script aims to be a precursor to fMRIprep
-
Any missing dependencies are installed as needed.
-
pydicom.pyis a script to probe dicoms and to get useful information (this step is not necessary but the information is useful to refer to later)
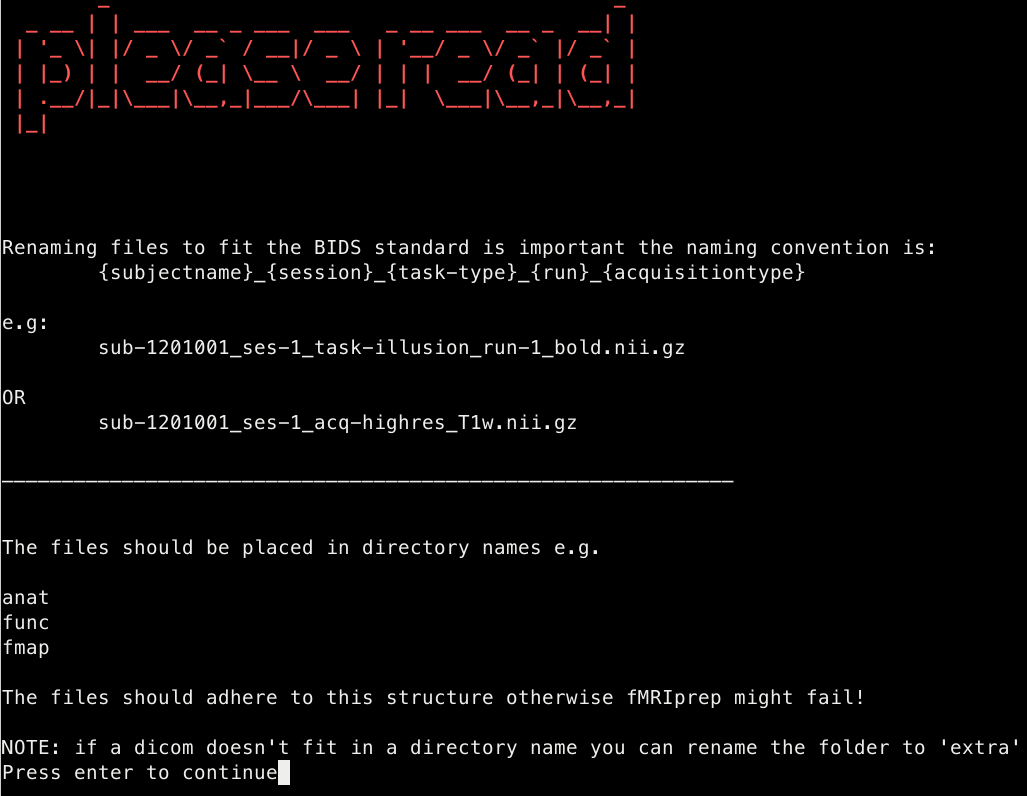
- Useful information is provided to the user about the BIDS format structure before the renaming takes place
- The user is able to change the names of the nifti files (this is automatically applied to the accompanying
.json)
-
Clone repo
git clone https://github.com/adam-coates/dicom2bids.git -
Ensure that dcm2niix is installed
-
The python module
pydicomandjqare automatically installed if not already found
-
Point to DICOM and BIDS directories
-
Select the DICOM folder
2a. Optionally select sub folder (e.g. ses-1/ses-*)
-
Rename the BIDS subject name (the script tries to derive the subject name itself although this is wrong and DICOM folders names vary)
-
Pydicom (or dcmunpack) will scan the DICOM directory and output a useful
.txtfile about the DICOM name and path -
DICOMS are roughly unpacked into the subject folder
-
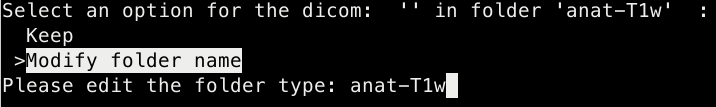
Keep the name of the nifti/ modify it or delete it
- If keeping the nifti name the script will ask the user whether to modify its folder name or keey it
-
If
fmapfolder present the script will try and create anIntendedFor.jsonfield entry and list thefuncfiles- An additional text file is created for the
IntendedForfield that is BIDS uri compliant
- An additional text file is created for the
always double check the conversion afterwards for any mistakes