- Document code : Done
- Clean notebooks : Done
- Make ReadMe : Almost Done
- Add the code for individual differences : WIP
- Clean code for glomeruli level metrics : WIP
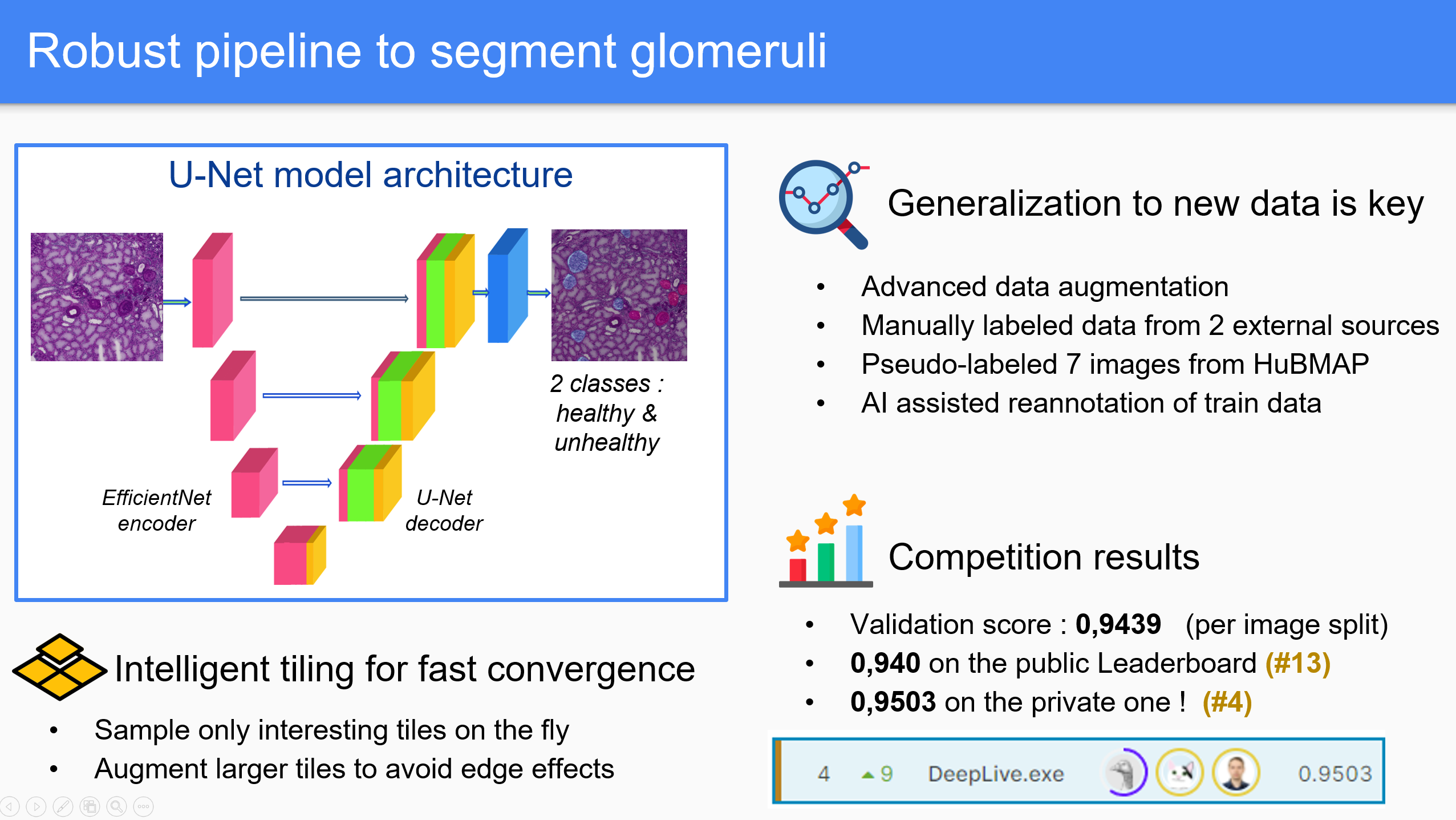
Our approach is built on understanding the challenges behind the data. Our main contribution is the consideration of the link between healthy glomeruli and unhealthy ones by predicting both into two different classes. We incorporate several external datasets in our pipeline and manually annotated the two classes. Our model architecture is relatively simple, and the pipeline can be easily transferred to other tasks.
You can read more about our solution here. A more concise write-up is also available here.
The main branch contains a cleaned and simplified version of our pipeline, that is enough to reproduce our solution.
Our pipeline achieves highly competitive performance on the task, because of the following aspects :
- It allows for fast experimenting and results interpretation:
- Pre-computation of resized images and masks of the desired size for short dataset preparation times
- Uses half-precision for faster training
- Interactive visualization of predicted masks in notebooks
- Glomeruli level metrics and confidence to understand the flaws of the model [TODO : Add code]
- It uses intelligent tiling, compatible with every segmentation task on big images.
- We sample only interesting regions, using mask and tissue information
- Tiling is made on the fly, on previously resized images for efficiency.
- Augmentations are made on slightly bigger tiles to get rid of side effects
- It is adapted to the specificity of the problem, to better tackle its complexity :
- We added another class to the problem : unhealthy glomeruli
- We manually annotated external data, as well as missing masks in the training data using model feedback.
- Aggressive augmentations help the model generalize well to quality issues in the test data
For the following reasons, our code is convenient to use, especially for researchers :
- It is only based on commonly used and reliable libraries:
- PyTorch
- Albumentations for augmentations
- Segmentations Models PyTorch for modeling
- It is easily re-usable:
- It is documented and formatted
- It includes best-practices from top Kagglers, who also have experience in research and in the industry
- It is (relatively) low level, which means one can independently use each brick of our pipeline in their code
- We applied our pipeline to keratinocytes segmentation in LC-OCT and quickly achieved good results. See here for more information.
-
Clone the repository
-
[TODO : Requirements]
-
Download the data :
- Put the competition data from Kaggle in the
inputfolder - Put the extra
Dataset Aimages from data.mendeley.com in theinput/extra/folder. - Put the two additional images from the HubMAP portal in the
input/test/folder. - You can download pseudo labels on Kaggle
- We also provide our trained model weights on Kaggle
- Put the competition data from Kaggle in the
-
Prepare the data :
- Extract the hand labels using
notebooks/Json to Mask.ipynb:- Use the
ADD_FCandONLY_FCparameters to generate labels for the healthy and unhealthy classes. - Use the
SAVE_TIFFparameter to save the external data as tiff files of half resolution. - Use the
PLOTparameter to visualize the masks. - Use the
SAVEparameter to save the masks as rle.
- Use the
- Create lower resolution masks and images using
notebooks/Image downscaling.ipynb:- Use the
FACTORparameter to specify the downscaling factor. We recommend generating data of downscaling 2 and 4. - For training data, we save extra time by also computing downscaling rles. Use the
NAMEparameter to specify which rle to downscale. Make sure to run the script for all the dataframes you want to use. - It is only require to save the downscaled images once, use the
SAVE_IMGparameters to this extent.
- Use the
- The process is a bit time-consuming, but only requires to be done once. This allows for faster experimenting : loading and downscaling the images when building the dataset takes a while, so we don't want to do it every time.
- Extract the hand labels using
-
Train models using
notebooks/Training.ipynb- Use the
DEBUGparameter to launch the code in debug mode (single fold, no logging) - Specify the training parameters in the
Configclass. Feel free to experiment with the parameters, here are the main ones :tile_size: Tile sizereduce_factor: Downscaling factoron_spot_sampling: Probability to accept a random tile with in the datasetoverlap_factor: Tile overlapping during inferenceselected_folds: Folds to run computations for.encoder: Encoder as defined in Segmentation Models PyTorchdecoder: Decoders from Segmentation Models PyTorchnum_classes: Number of classes. Keep it at 2 to use the healthy and unhealthy classesloss: Loss function. We use the BCE but the lovasz is also interestingoptimizer: Optimizer namebatch_size: Training batch size, adapt theBATCH_SIZESdictionary to your gpuval_bs: Validation batch sizeepochs: Number of training epochsiter_per_epoch: Number of tiles to use per epochlr: Learning rate. Will be decayed linearlywarmup_prop: Proportion of steps to use for learning rate warmupmix_proba: Probability to apply MixUp withmix_alpha: Alpha parameter for MixUpuse_pl: Probability to sample a tile from the pseudo-labeled imagesuse_external: Probability to sample a tile from the external imagespl_path: Path to pseudo labels generated bynotebooks/Inference_test.ipynbextra_path: Path to extra labels generated bynotebooks/Json to Mask.ipynb(should not be changed)rle_path: Path to train labels downscaled bynotebooks/Image downscaling.ipynb(should not be changed)
- Use the
-
Validate models with
notebooks/Inference.ipynb:- Use the
log_folderparameter to specify the experiment. - Use the
use_ttaparameter to specify whether to use test time augmentations. - Use the
saveparameter to indicate whether to save predictions. - Use the
save_all_ttaparameter to save predictions for each tta (takes a lot of disk space). - Use the
global_thresholdparameter to tweak the threshold.
- Use the
-
Generate pseudo-labels with
notebooks/Inference Test.ipynb:- Use the
log_folderparameter to specify the experiment. - Use the
use_ttaparameter to speciy whether to use test time augmentations. - Use the
saveparameter to indicate whether to save predictions.
- Use the
-
Visualize predictions :
notebooks/Visualize Predictions.ipynb- Works to visualize predictions from the two previous notebooks, but also from a submission file.
- Specify the
name,log_folderandsubparameters according to what you want to plot.
If you wish to dive into the code, the repository naming should be straight-forward. Each function is documented. The structure is the following :
code
├── data
│ ├── dataset.py # Torch datasets
│ └── transforms.py # Augmentations
├── inference
│ ├── main_test.py # Inference for the test data
│ └── main.py # Inference for the train data
├── model_zoo
│ └── models.py # Model definition
├── training
│ ├── lovasz.py # Lovasz loss implementation
│ ├── main.py # k-fold and training main functions
│ ├── meter.py # Meter for evaluation during training
│ ├── mix.py # CutMix and MixUp
│ ├── optim.py # Losses and optimizer handling
│ ├── predict.py # Functions for prediction
│ └── train.py # Fitting a model
├── utils
│ ├── logger.py # Logging utils
│ ├── metrics.py # Metrics for the competition
│ ├── plots.py # Plotting utils
│ ├── rle.py # RLE encoding utils
│ └── torch.py # Torch utils
└── params.py # Main parameters