Multi-group Shared Subspace Covariance Estimation
Reference: Shared Subspace Models for Multi-Group Covariance Estimation (JMLR, 2019)
Install mgCov using devtools:
# install.packages("devtools")
devtools::install_github("afranks86/mgCov")We’ll demonstrate the use of mgCov with a gene expression dataset from
patients with multiple subtypes of acute lymphoblastic leukemia from
Yeoh et al
(2002).
For simplicity we include a subset of 1000 genes. The data includes
expression levels on 327 split across 7 leukemia subtypes.
library(mgCov)
data(leukemia)
sapply(data_list, function(x) nrow(x))
#> BCR-ABL E2A-PBX1 Hyperdip50 MLL OTHERS T-ALL
#> 15 27 64 20 79 43
#> TEL-AML1
#> 79S <- getRank(data_list)
Vinit <- mgCov::subspaceInit(data_list, S)
EMFit <- subspaceEM(data_list, S=S)
#> [1] "Reached maximum iterations in line search."
Vfit <- EMFit$V ## inferred basis for the shared subspaceNow run (conditional) Bayesian covariance estimation using the inferred subspace.
samples <- fitBayesianSpike(V=Vfit, Ylist=data_list,
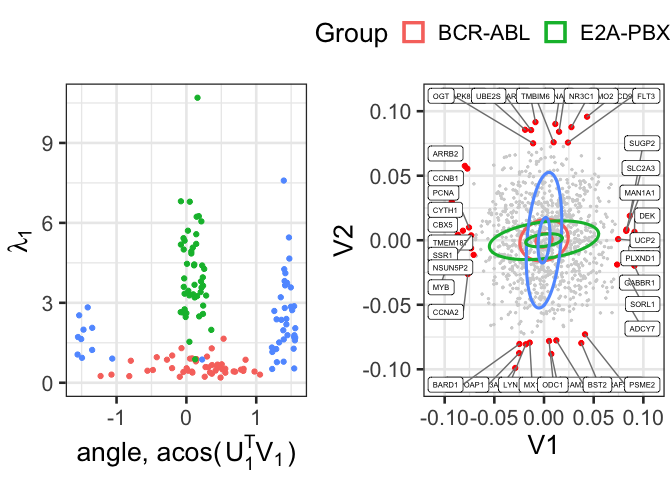
niters=1000, nskip=10, verbose=FALSE)Let’s compare the gene expression covariance matrices of E2A-PBX1 to MLL.
groups_to_plot = c(1, 2, 4)
names(groups_to_plot) <- names(data_list)[groups_to_plot]
create_plots(V=Vfit, samples, group1=2, group2=4, to_plot = groups_to_plot, view=c(1, 2))We can compare the same groups on a different two dimensional subspace.
By setting view to …
This is analogous to looking at the 3rd and 4th principal components in a standard PCA.
create_plots(V=Vfit, samples, group1=2, group2=4, to_plot = groups_to_plot, view=c(3, 4))
#> Warning: Removed 1 rows containing missing values (geom_label_repel).We can compare different groups
create_plots(V=Vfit, samples, group1=2, group2=4, to_plot = groups_to_plot, view=c(3, 4))
#> Warning: Removed 1 rows containing missing values (geom_label_repel).TO DO