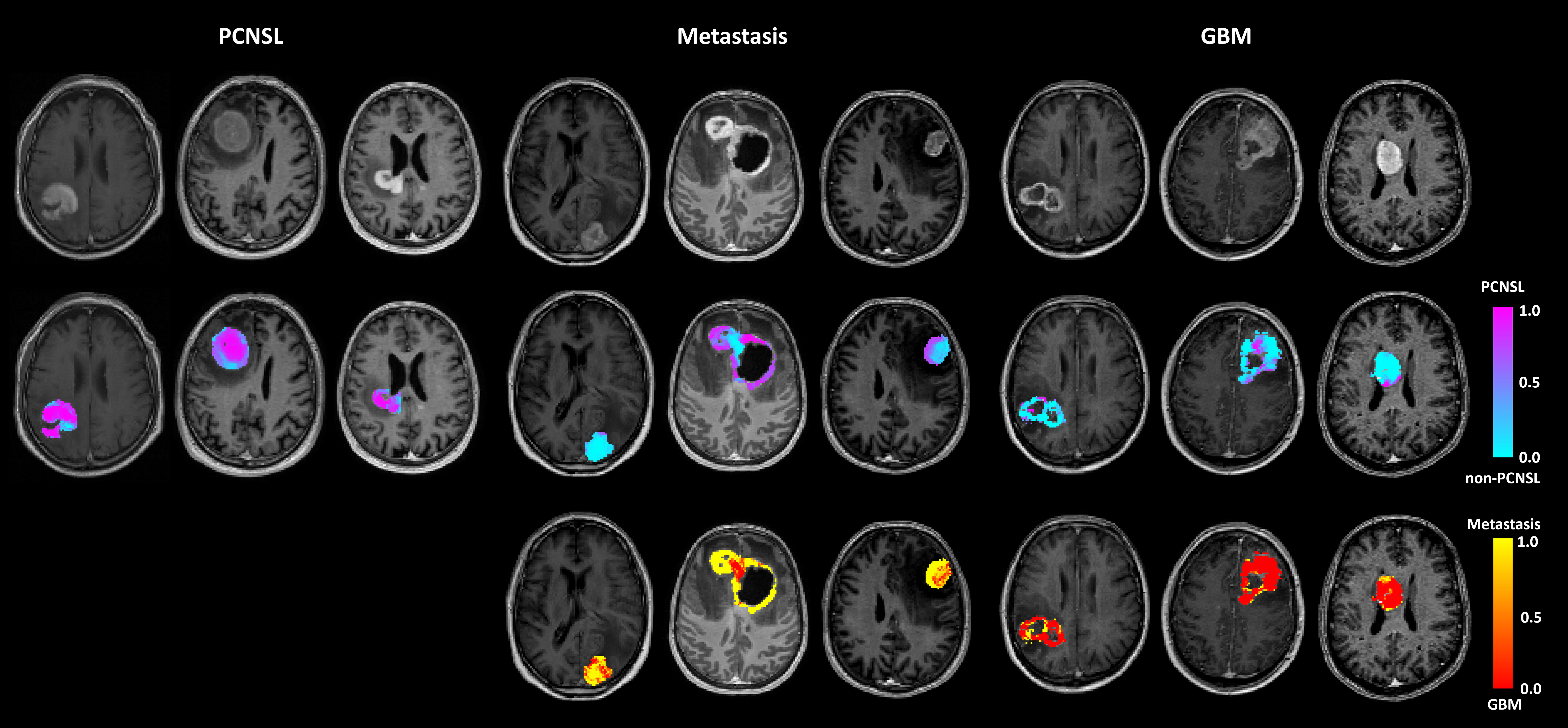
The app uses a pretrained CNN classifier scheme for voxel-wise classification of enhancing brain tumours into P-CNS-lymphoma, glioblastoma or brain metastasis, from dynamic susceptibility contrast (DSC) MRI data.
This is the code repository of the brain tumour classification pipeline described in the work An accessible deep learning tool for voxel-wise classification of brain malignancies from perfusion MRI by Raquel Perez-Lopez and colleagues from the Vall d'Hebron Institute of Oncology (VHIO), Barcelona, Spain.
The app will be running in our servers on link.
Visit us here.
- Python 3.8
- Python packages listed in
requirements.txt - Optionally, to handle compressed DICOMs: pydicom (2.1.2 tested), gdcm (2.8.9 tested)
- If used without graphical interface in Linux systems, xvfb-run is needed (
apt-get install xvfb-run)
Download or clone the app repo in a directory, this will create the folder /discern-app:
git clone https://github.com/radiomicsgroup/discern-app
Though not necessary, you may create a new python/conda environment:
conda create --name myenv python=3.8
conda activate myenv
Install python packages specified in the file requirements.txt from the discern-app folder:
pip install -r requirements.txt
Optionally:
pip install pydicom==2.1.2
pip install gdcm==2.8.9
Download Slicer from above links into, e.g. /slicer
Download dcm2niix from above links into, e.g. /dcm2niix
In settings.py, place the path of the Slicer executable as: SLICER_EXEC="/slicer/Slicer"
In settings.py, place the path of the dcm2niix executable as: DCM2NIIX="/dcm2niix/dcm2niix"
To run the pipeline, call the run.py file (full path python /discern-app/run.py):
python run.py --p_dsc /path_to_DSC_image --p_t1 /path_to_T1_image --p_output /desired_output_path
Input DSC and T1wCE volumes may be in DICOM, Nifti or NRRD formats. Segmentation mask files can be provided instead of the T1 volume.
See all the options in the docstring help of run.py.
- Preprint:
Raquel Perez-Lopez et al., An accessible deep learning tool for voxel-wise classification of brain malignancies from perfusion MRI
- Github:
Please, see license.txt
- The use of any software provided by VHIO as part of its services, from now on referred to as the services, is subject to the terms and conditions of the third party license agreements applicable to the relevant software as made available through the website [https://github.com/radiomicsgroup/discern-app].
- The services shall be used solely for research activities that comply with all applicable laws and regulations.
- Users shall keep all user IDs, passwords and other means of access to the services account within its possession or control and shall keep those confidential and secure from unauthorised access or use.
- If user becomes aware of any unauthorized use of a password or the services account, user will notify the VHIO team as promptly as possible through the provided channels.
- User will not use the services to identify the individuals who are data subjects.
- Do NOT use any of the services if the safety of data subjects depends directly on the uninterrupted, and error free availability of the services at all times, or the compatibility or operation of the services with all hardware and software configurations.
- Within the context of a study, the principal investigator is responsible for:
- Verification of the identity of users and delegates;
- Order assignment and withdrawal of access authorizations for users and delegates;
- Informing users and delegates about the responsible handling of study data.
- Within the context of a study, users may only use the services as per instructions of the study’s principal investigator, specifically:
- Data upload and download to and from the services shall only be executed in accordance with the study conditions and with the consent of the principal investigator.