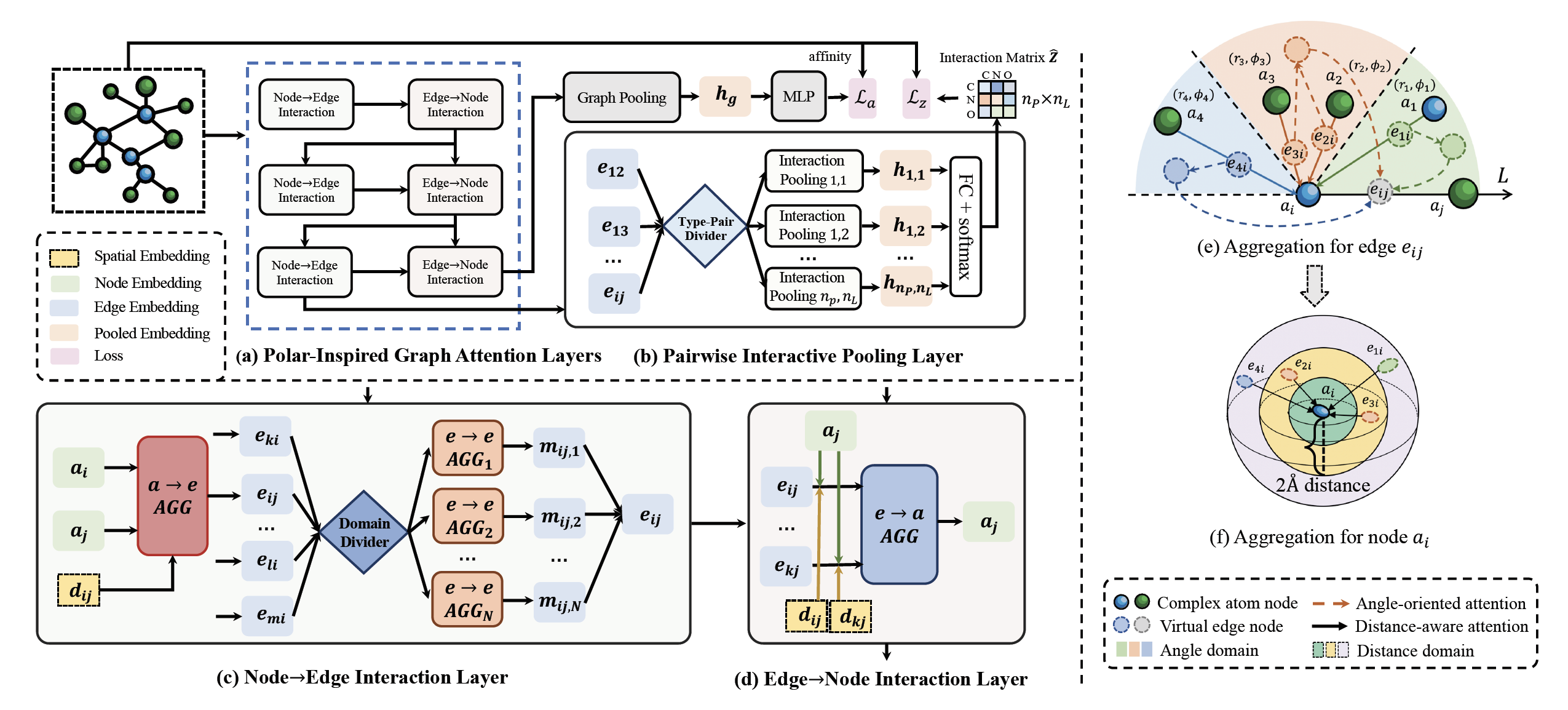
Source code for KDD 2021 paper: "Structure-aware Interactive Graph Neural Networks for the Prediction of Protein-Ligand Binding Affinity".
- python >= 3.8
- paddlepaddle >= 2.1.0
- pgl >= 2.1.4
- openbabel == 3.1.1 (optional, only for preprocessing)
The PDBbind dataset can be downloaded here.
The CSAR-HiQ dataset can be downloaded here.
You may need to use the UCSF Chimera tool to convert the PDB-format files into MOL2-format files for feature extraction at first.
Alternatively, we also provided a dropbox link for downloading PDBbind and CSAR-HiQ datasets.
The downloaded dataset should be preprocessed to obtain features and spatial coordinates:
python preprocess_pdbbind.py --data_path_core YOUR_DATASET_PATH --data_path_refined YOUR_DATASET_PATH --dataset_name pdbbind2016 --output_path YOUR_OUTPUT_PATH --cutoff 5
The parameter cutoff is the threshold of cutoff distance between atoms.
You can also use the processed data from this link. Before training the model, please put the downloaded files into the directory (./data/).
To train the model, you can run this command:
python train.py --cuda YOUR_DEVICE --model_dir MODEL_PATH_TO_SAVE --dataset pdbbind2016 --cut_dist 5 --num_angle 6
If you find our work is helpful in your research, please consider citing our paper:
@inproceedings{li2021structure,
title={Structure-aware Interactive Graph Neural Networks for the Prediction of Protein-Ligand Binding Affinity},
author={Li, Shuangli and Zhou, Jingbo and Xu, Tong and Huang, Liang and Wang, Fan and Xiong, Haoyi and Huang, Weili and Dou, Dejing and Xiong, Hui},
booktitle={Proceedings of the 27th ACM SIGKDD Conference on Knowledge Discovery \& Data Mining},
pages={975--985},
year={2021}
}