Problem related to STRIP-AI competition, to classify the blood clot origins in ischemic stroke. Using whole slide digital pathology images to differentiate between the two major acute ischemic stroke (AIS) etiology subtypes: cardiac and large artery atherosclerosis (CE - LAA).
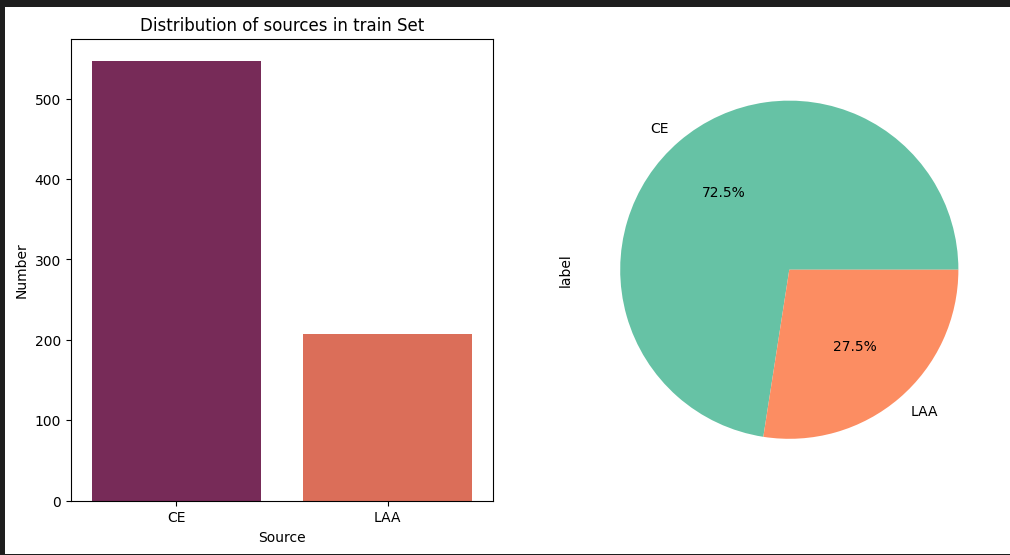
The data comprises 754 high-resolution whole-slide digital pathology images in TIF format, categorizing stroke origin as either CE or LAA, but have size variations and the distribution of the classes is not balanced that need careful handling.
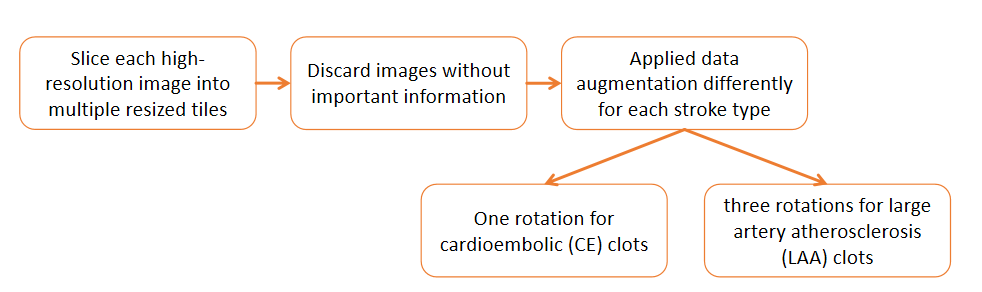
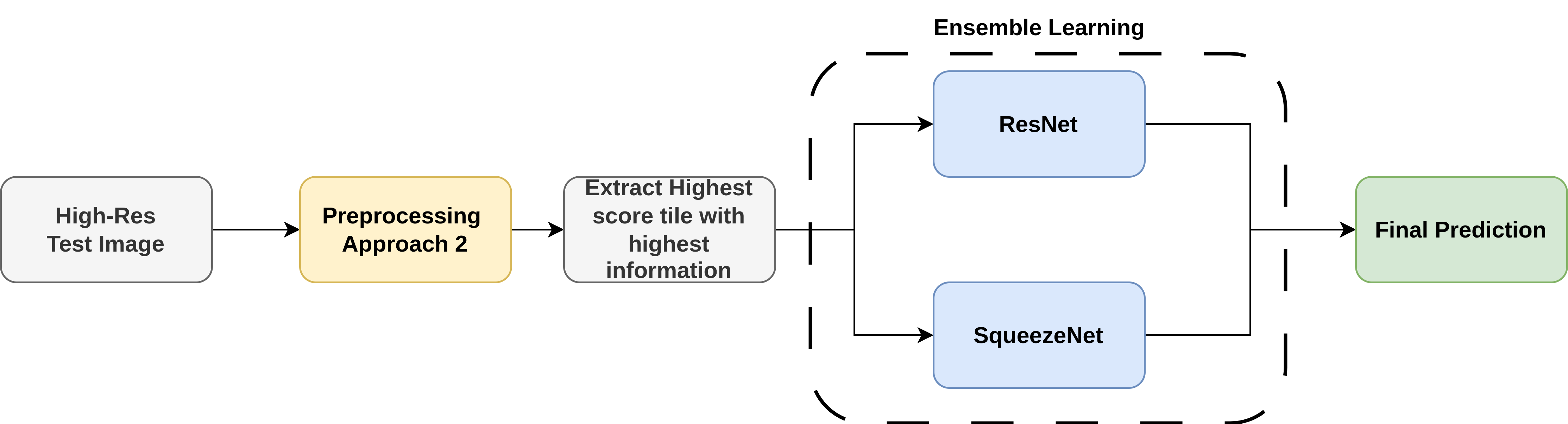
To tackle the challenges of limited, imbalanced, and variable-sized stroke clot images, we leveraged data augmentation. effectively balancing the dataset and boosting training data volume( 4313 final images).Ensemble learning between ResNet and SqueezeNet
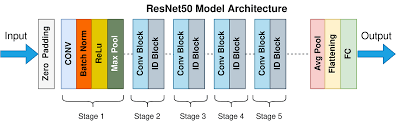
- ResNet50 Model:
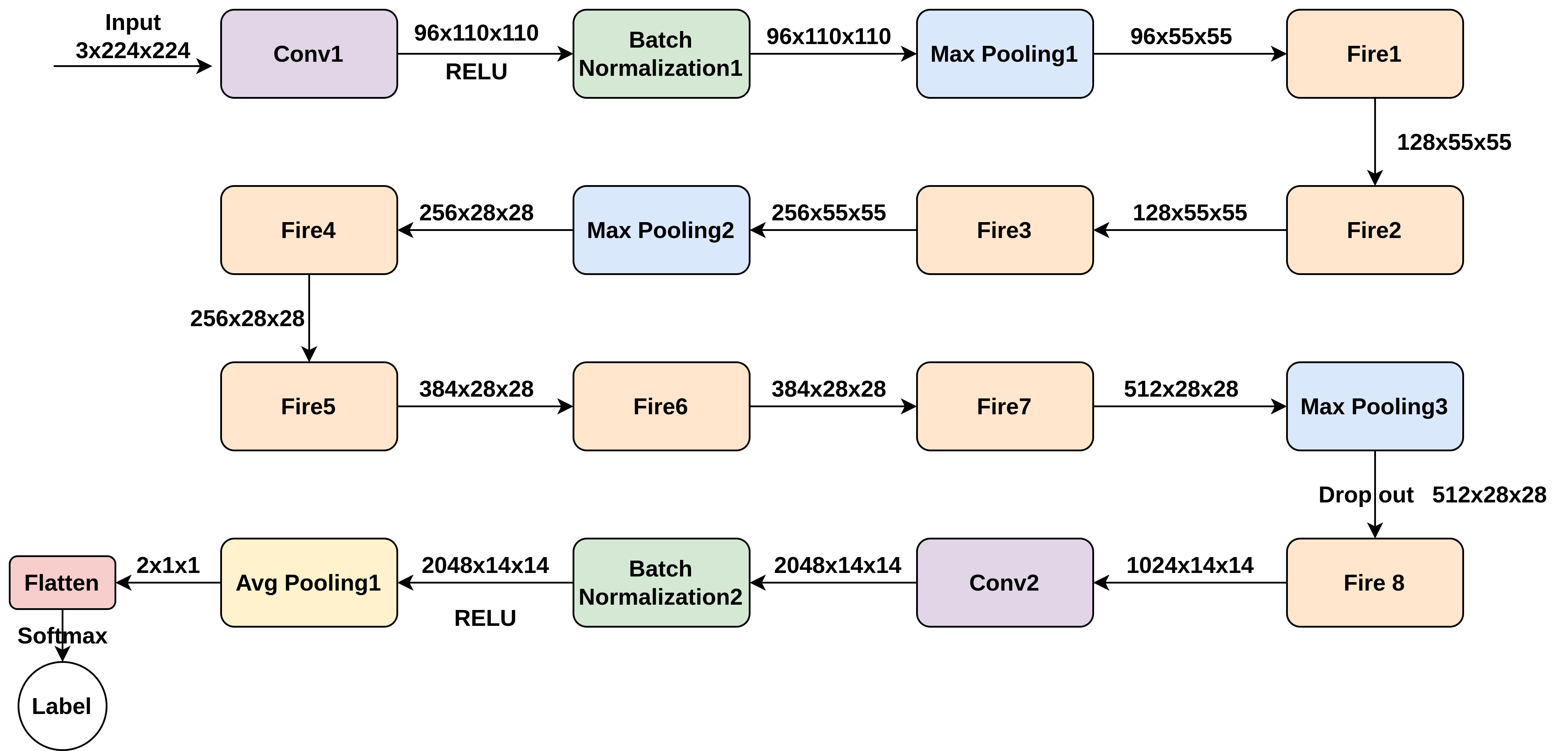
- SqueezeNet Model:
Test Pipeline for one test image:
In This part, we will see some results of each model we tried.
- Confusion Matrix
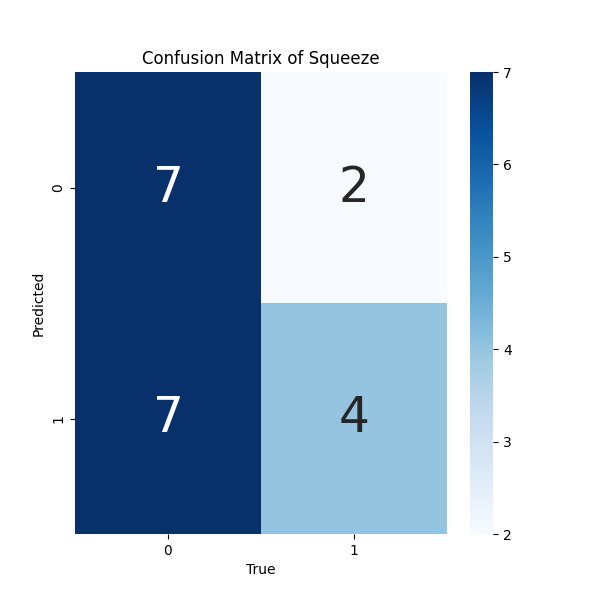
- Confusion Matrix for results of SqueezeNet Model:
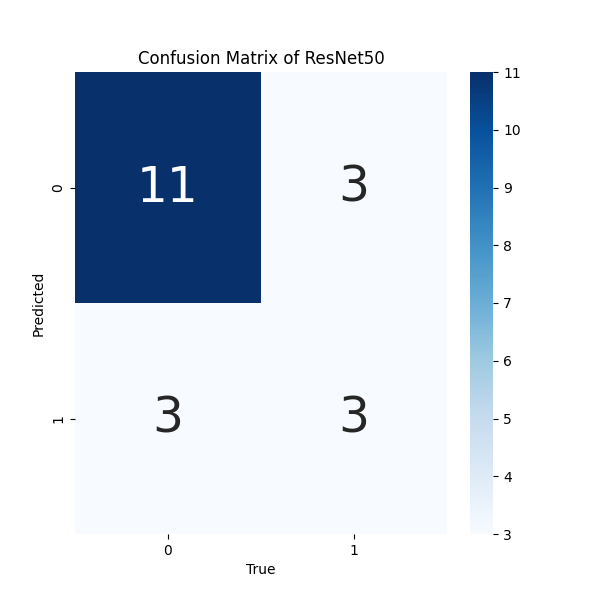
- Confusion Matrix for results of ResNet50 Model:
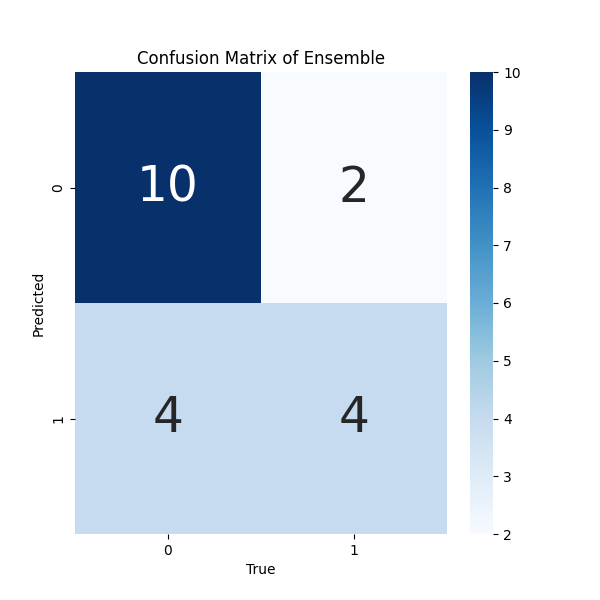
- Confusion Matrix for results of Ensemble Model:
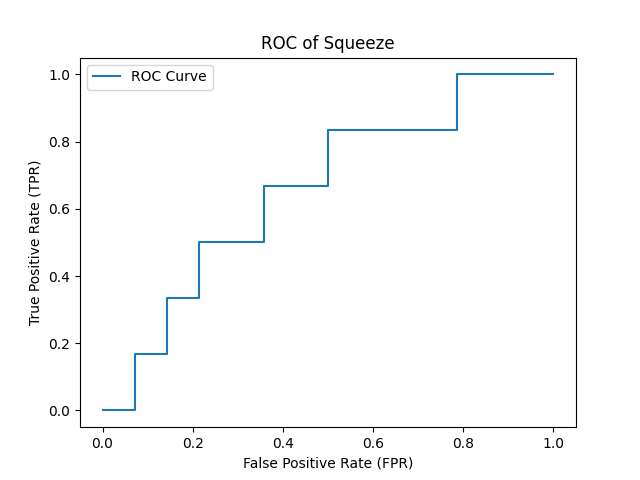
- ROC Curve for results of SqueezeNet Model with AUC = 0.65 :
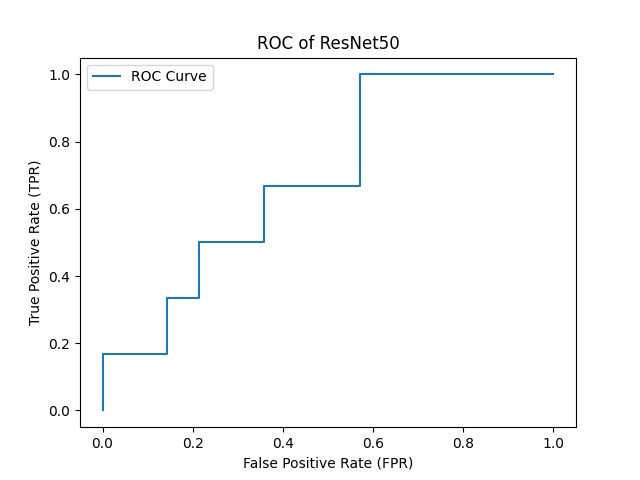
- ROC Curve for results of ResNet50 Model with AUC = 0.69 :
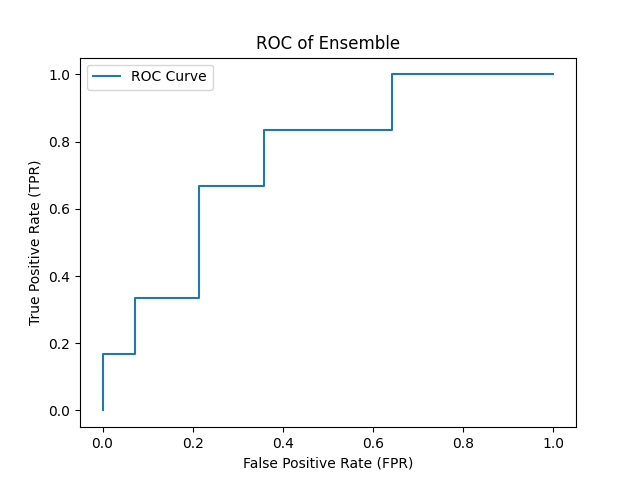
- ROC Curve for results of Ensemble Model with AUC = 0.75 :
| Metric | Score |
|---|---|
| Precision | 0.833 |
| Recall | 0.714 |
| F1 Score | 0.769 |
| Accuracy | 0.700 |
| Specificity | 0.666 |
main
├─ Config
├─ data: dir containing 2 data samples
│ ├─ raw : original data
│ ├─ processed: Images Tiles
│ └─ processed-after-removing-low-score-imgs: Tiles after filtering
├─ models: containing models checkpoints
├─ notebooks
│ ├─ Data Exploring.ipynb
│ ├─ Data Preprocessing Approach 1.ipynb
│ ├─ Data Preprocessing Approach 2.ipynb
│ ├─ EfficientNet_training_kaggle.ipynb
│ ├─ RestNet5_training_kaggle.ipynb
│ ├─ SqueezeNet_training_kaggle.ipynb
│ └─ test-evaluation.ipynb
├─ reports
│ ├─ Paper.pdf
| └─ figures
├─ src
│ ├─ preprocessing
│ │ ├─ preprocessing_approach_1.py
│ │ └─ preprocessing_approach_2.py
| ├─ dataloaders
│ │ ├─ Dataloader.py
│ │ └─ Dataloader_with_aug.py
│ └─ models
│ ├─ EfficientNet.py
│ ├─ ResNet50.py
│ ├─ squeezeNet.py
│ └─ helpers
│ └─ FireModule.py
└─ README.md
Currently you can run and test project through these notebooks on Kaggle using competition data :
main
└─ notebooks
├─ Data Preprocessing Approach 2.ipynb
├─ RestNet5_training_kaggle.ipynb
├─ SqueezeNet_training_kaggle.ipynb
└─ test-evaluation.ipynb
Also, You can find models checkpoints on
First Semester - Artificial Neural Networks in Medicine (SBE4025) class project created by:
| Team Members' Names | Code |
|---|---|
| Ahmed Hassan | 9202076 |
| Habiba Fathallah | 9202458 |
| Rawan Mohamed | 9202559 |
| Romaisaa Saad | 9202564 |
- Dr. Inas Yassine & Eng. Merna Biabers All rights reserved © 2024 to Team 3 - Systems & Biomedical Engineering, Cairo University (Class 2024)