Improving the Taxonomy of Fossil Pollen using Convolutional Neural Networks and Superresolution Microscopy
[PNAS paper] [NSF News]
Models and data are hosted in google drive and Illinois Databank (to be updated).
Authors: Ingrid C. Romero, Shu Kong, Charless C. Fowlkes, Michael A. Urban, Carlos Jaramillo, Francisca Oboh-Ikuenobe, Carlos D'Apolito, Surangi W. Punyasena
@article{deepPollen,
title={Improving the Taxonomy of Fossil Pollen using Convolutional Neural Networks and Superresolution Microscopy},
author={Romero, Ingrid C. and Kong, Shu and Fowlkes, Charless C. and Jaramillo, Carlos and Urban, Michael A. and Oboh-Ikuenobe, Francisca and D’Apolito, Carlos and Punyasena, Surangi W.},
journal={Proceedings of the National Academy of Sciences (PNAS)},
year={2020}
}
Last edited: Sep. 16, 2020 (this repository is being actively updated)
Keywords: Airyscan microscopy, automated classification, Detarioideae, machine learning, palynology.
Significance Statement: By combining optical superresolution with deep learning classification methods, we demonstrate that it is possible to taxonomically separate pollen grains that appear morphologically similar under light microscopy. This new approach improves the taxonomic resolution of pollen identifications and greatly enhances the use of pollen data in ecological and evolutionary research.
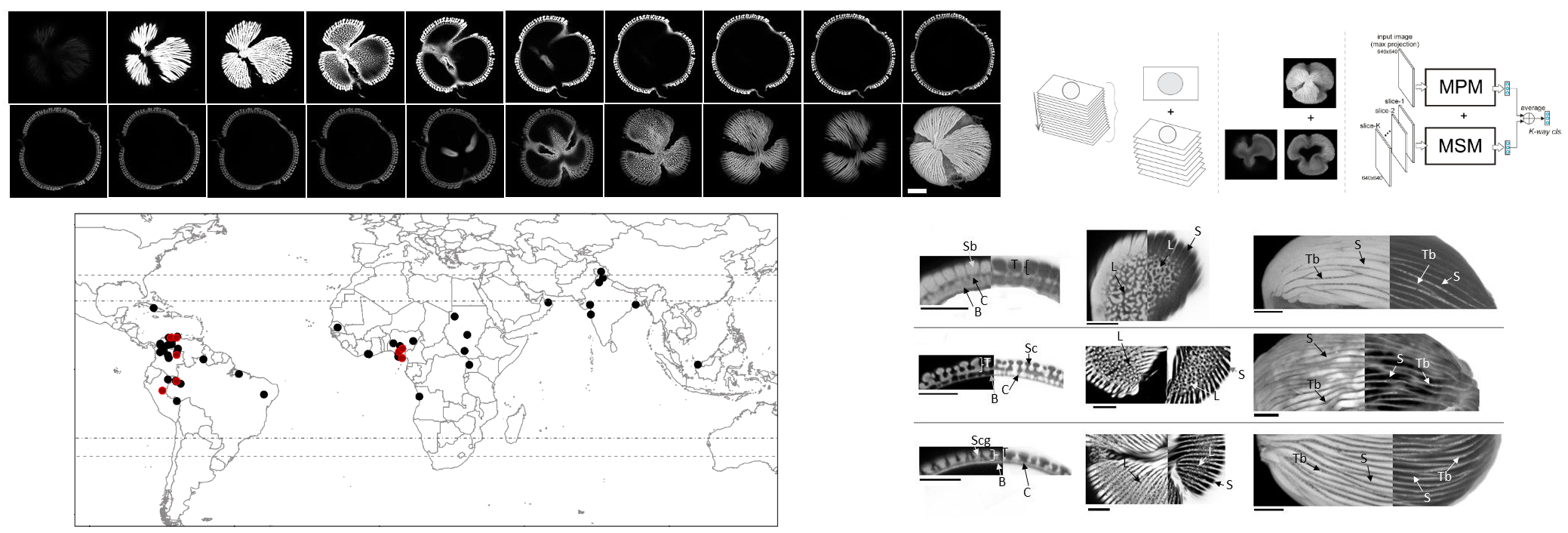
Taxonomic resolution is a major challenge in palynology, largely limiting the ecological and evolutionary interpretations possible with deep-time fossil pollen data. We present a new approach for fossil pollen analysis that uses optical superresolution microscopy and machine learning to create a quantitative and higher throughput workflow for producing palynological identifications and hypotheses of biological affinity. We developed three convolutional neural network (CNN) classification models: maximum projection (MPM), multi-slice (MSM), and fused (FM). We trained the models on the pollen of 16 genera of the legume tribe Amherstieae, and then used these models to constrain the biological classifications of 48 fossil Striatopollis specimens from the Paleocene, Eocene, and Miocene of western Africa and northern South America. All models achieved average accuracies of 83 - 90% in the classification of the extant genera, and the majority of fossil identifications (86%) showed consensus among at least two of the three models. Our fossil identifications support the paleobiogeographic hypothesis that Amherstieae originated in Paleocene Africa and dispersed to South America during the Paleocene-Eocene Thermal Maximum (56 Ma). They also raise the possibility that at least three Amherstieae genera (Crudia, Berlinia, and Anthonotha) may have diverged earlier in the Cenozoic than predicted by molecular phylogenies.
We demonstrate that combining optical superresolution imaging with deep learning classification methods increases the speed and accuracy of assessing the biological affinities of fossil pollen taxa. We show that it is possible to taxonomically separate pollen grains that appear morphologically similar under standard light microscopy based on nanoscale variation in pollen shape, texture, and wall structure. Using a single pollen morphospecies, Striatopollis catatumbus, we show that nanoscale morphological variation within the fossil taxon coincides with paleobiogeographic distributions. This new approach improves the taxonomic resolution of fossil pollen identifications and greatly enhances the use of pollen data in ecological and evolutionary research.
There are two folders in thie repository.
-
Data Processing contains a set of processing steps such as segmentation of foreground pollen grains, rotating the grain to upright canonical viewpoint, max-projection, etc. Please go to this folder and refer to the readme file for details.
-
Training contains scripts that train the three models described in the paper. There are a few data samples that the code can run on them smoothly.
-
Testing shows how we run the trained models during testing. The three types of trained models can be downloaded in the google drive. Please put them under "./demo_step003_training/models" to run them.
contact: Shu Kong