This project would aid the researchers in evaluating the DNA damage of cells under stress. Helps to dentifying and quantifying the nuclei (cell).
- Python - v3.10.8
- cv2 - 4.6.0
- matplotlib - 3.6.2
- pandas - 1.5.2
- skimage - 0.19.3
Clone the repository, install the dependencies and get started right away.
$ git clone git@github.com:aishstha/Quantify-nuclei.git
$ cd Quantify-nuclei
$ ./segment_script.py <Input-image-file-path>
Example :
$ ./segment_script.py "Data/IXMtest_C05_s7_w1F71963FB-8F29-41CB-A5F5-07CB9584BBC5.tif"
You can manually Install packages mentioned above (cv2, numpy, matplotlib, pandas, sys, skimage).
OR
Use the conda environment file of this project. For more information, check environment.yml
Use the terminal or an Anaconda Prompt for the following steps:
- Create the environment from the environment.yml file:
$ conda env create -f environment.yml
-
Activate the new environment:
conda activate segmentation -
Verify that the new environment was installed correctly:
$ conda env list
After running the script, check data_to_excel.xlsx file for stats and output.jpg for visual result. The output.jpg file has the labeling in the cells, the information about each cell is according to the label in excel file.
| Input | Output |
|---|---|
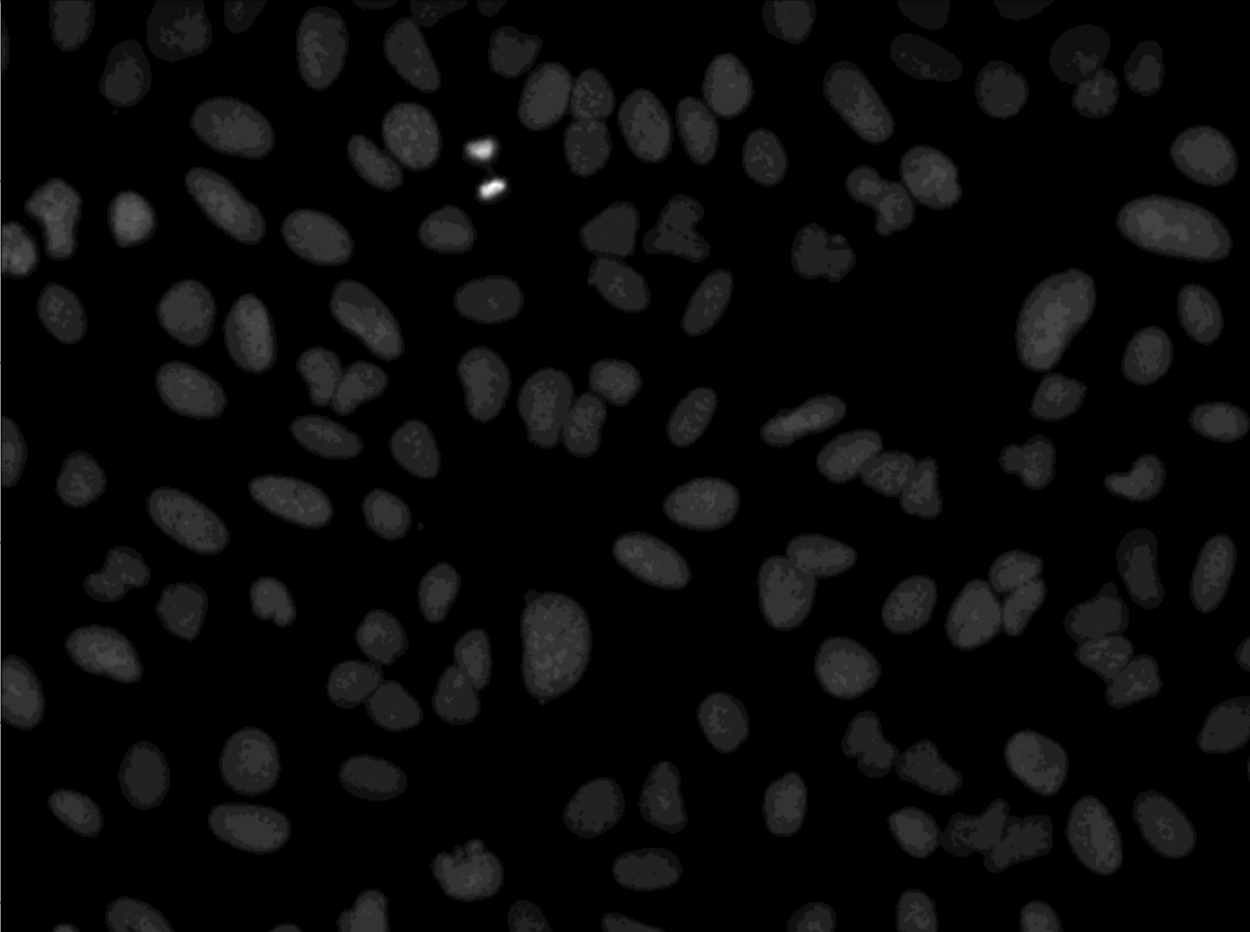 |
 |