Dataplug is a client-side only, extensible, Python framework with the goal of enabling efficient data partitioning of unstructured scientific data stored in object storage (like Amazon S3) for elastic workloads in the Cloud
-
Dataplug provides a plug-in interface to enable data partitioning to a multitude of scientific data types, from a variety of domains, stored in S3. Currently, Dataplug supports the following data types:
-
Dataplug follows a read-only cloud-aware pre-processing approach to enable on-the-fly dynamic partitioning of scientific unstructured data.
- It is cloud-aware because it specifically targets cold raw data residing in huge repositories in object storage (e.g. Amazon S3). S3 allows partial reads at high bandwidth by using HTTP GET Byte-range requests. Dataplug builds indexes around this premise to enable parallel chunked access to unstructured data. It compensates high latency of object storage with many parallel reads at high bandwidth.
- It is Read-only because, in object storage, objects are immutable. Thus, pre-processing is read-only, meaning that index and metadata are stored decoupled from data, in another object. Raw cold data is kept as-is in storage. This voids to re-write the entire dataset for partitioning. Since indexes are several orders of magnitude smaller, the data movements are considerably reduced.
-
Dataplug allows re-partitioning a dataset at zero-cost.
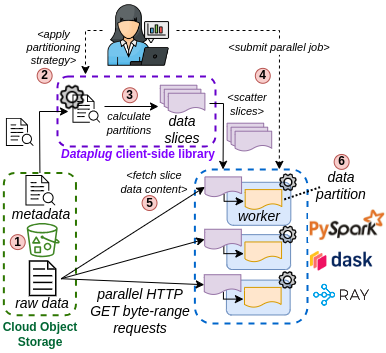
- Dataplug introduces the concept of data slicing. A data slice is a lazily-evaluated partition of a pre-processed dataset in its raw form, present in object storage (1).
- Users can perform different partitioning strategies (2) on the same dataset without actually moving data around (3).
- Data slices are serializable, and can be sent to remote workers using any Python-compatible distributed computing environment (4) (e.g. PySpark, Dask or Ray).
- Data slices are evaluated at the moment of accessing the data, and not before (5). This allows many remote workers to perform many HTTP GET Byte-range requests in parallel onto the raw dataset, exploiting S3's high bandwidth capabilities.
-
Dataplug is only available through GitHub. You can use
pipto install it directly from the repository:pip install git+https://github.com/CLOUDLAB-URV/dataplug
from dataplug import CloudObject
from dataplug.formats.genomics.fastq import FASTQGZip, partition_reads_batches
# Assign FASTQGZip data type for object in s3://genomics/SRR6052133_1.fastq.gz
co = CloudObject.from_s3(FASTQGZip, "s3://genomics/SRR6052133_1.fastq.gz")
# Data must be pre-processed first ==> This only needs to be done once per dataset
# Preprocessing will create reusable indexes to repartition
# the data many times in many chunk sizes
# Dataplug leverages joblib to deploy preprocessing jobs
co.preprocess(parallel_config={"backend": "dask"})
# Partition the FASTQGZip object into 200 chunks
# This does not move data around, it only creates data slices from the indexes
data_slices = co.partition(partition_reads_batches, num_batches=200)
def process_fastq_data(data_slice):
# Evaluate the data_slice, which will perform the
# actual HTTP GET requests to get the FASTQ partition data
fastq_reads = data_slice.get()
...
# Use Dask for deploying a parallel distributed job
import dask.bag as db
from dask.distributed import Client
client = Client()
# Create a Dask Bag from the data_slices list
dask_bag = db.from_sequence(data_slices)
# Apply the process_fastq_data function to each data slice
# Dask will serialize the data_slices and send them to the workers
dask_bag.map(process_fastq_data).compute()This project has been partially funded by the EU Horizon programme under grant agreements No. 101086248, No. 101092644, No. 101092646, No. 101093110.