Beta: this is still under development. Contact me if you need help. We'll be adding features and possibly making breaking changes in future releases.
Look for other our projects at https://github.com/RNA-Puzzles.
Magnus, Marcin. (2016). rna-pdb-tools. Zenodo. 10.5281/zenodo.60933
If you find the tools helpful, you can cite the repo using the doi above :-), you can donate via PayPal, and if you like the project, please "Star it", so it would be easier to find it for others and to make me happy that the toolbox useful not only for me.
A core library and a set of programs to run various Python functions related to work with PDB files of RNA structures.
The software is used by me in my servers NPDock (RNA/DNA-protein docking method, http://genesilico.pl/NPDock/) and SimRNAweb (RNA 3D structure prediction method, http://iimcb.genesilico.pl/SimRNAweb/) and mqapRNA (RNA 3D quality control, http://iimcb.genesilico.pl/mqapRNA/).
What is fun here?
rna-pdb-tools is a packages of shell utils that are using the common core library. You can also access functions of the library from your scripts.
A shell util:
$ rna_pdb_toolsx.py --is_pdb input/1I9V_A.pdb
True
$ rna_pdb_toolsx.py --is_pdb input/image.png
Falseor from a script
>>> from rna_pdb_tools_lib import *
>>> s = RNAStructure('input/1I9V_A.pdb')
>>> s.is_pdb()
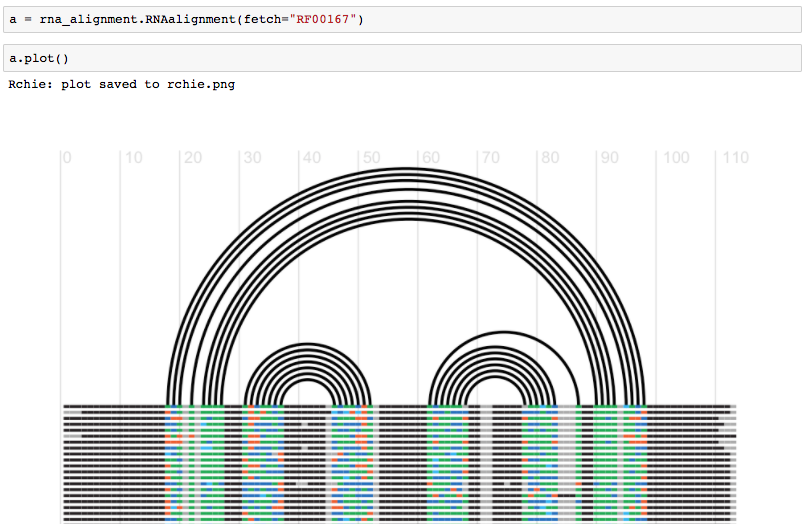
Trueor from another script (fetch an alignment and plot it)
Fig. See more https://github.com/mmagnus/rna-pdb-tools/blob/master/rna_pdb_tools/utils/rna_alignment/rna_alignment.ipynb
Take a tour http://mmagnus.github.io/rna-pdb-tools/#/ and/or read the doc rna-pdb-tools.rtfd.io/en/latest/.
Fig. rna_pdb_toolsx.py --get_rnapuzzle_ready *pdb --inplace
Take a tour http://mmagnus.github.io/rna-pdb-tools/#/
[mm] rna_pdb_tools$ git:(master) ✗ ./rna_pdb_toolsx.py -h
usage: rna_pdb_toolsx.py [-h] [--version] [-r] [--delete-anisou] [-c]
usage: rna_pdb_toolsx.py [-h] [--version] [-r] [-c] [--is_pdb] [--is_nmr]
[--un_nmr] [--orgmode] [--get_chain GET_CHAIN]
[--fetch] [--fetch_ba] [--get_seq] [--get_ss]
[--rosetta2generic] [--get_rnapuzzle_ready] [--rpr]
[--no_hr] [--renumber_residues]
[--dont_rename_chains] [--dont_fix_missing_atoms]
[--dont_report_missing_atoms] [--collapsed_view]
[--cv] [-v] [--replace_hetatm] [--inplace]
[--mutate MUTATE] [--edit EDIT]
[--replace-chain REPLACE_CHAIN] [--delete DELETE]
[--extract EXTRACT]
file [file ...]
rna_pdb_tools - a swiss army knife to manipulation of RNA pdb structures
Tricks:
for i in *pdb; do rna_pdb_toolsx.py --get_rnapuzzle_ready $i > ${i/.pdb/_rpr.pdb}; done
Usage::
$ for i in *pdb; do rna_pdb_toolsx.py --delete A:46-56 $i > ../rpr_rm_loop/$i ; done
$ rna_pdb_toolsx.py --get_seq *
# BujnickiLab_RNApuzzle14_n01bound
> A:1-61
# BujnickiLab_RNApuzzle14_n02bound
> A:1-61
CGUUAGCCCAGGAAACUGGGCGGAAGUAAGGCCCAUUGCACUCCGGGCCUGAAGCAACGCG
[...]
positional arguments:
file file
optional arguments:
-h, --help show this help message and exit
--version
-r, --report get report
--delete-anisou remove files with ANISOU records
-c, --clean get clean structure
--is_pdb check if a file is in the pdb format
--is_nmr check if a file is NMR-style multiple model pdb
--un_nmr Split NMR-style multiple model pdb files into
individual models [biopython]
--orgmode get a structure in org-mode format <sick!>
--get_chain GET_CHAIN
get chain, .e.g A
--fetch fetch file from the PDB db
--fetch_ba fetch biological assembly from the PDB db
--get_seq get seq
--get_ss get secondary structure
--rosetta2generic convert ROSETTA-like format to a generic pdb
--get_rnapuzzle_ready
get RNApuzzle ready (keep only standard atoms).Be
default it does not renumber residues, use
--renumber_residues [requires biopython]
--rpr alias to get_rnapuzzle ready)
--no_hr do not insert the header into files
--renumber_residues by defult is false
--dont_rename_chains used only with --get_rnapuzzle_ready. By default
--get_rnapuzzle_ready rename chains from ABC.. to stop
behavior switch on this option
--dont_fix_missing_atoms
used only with --get_rnapuzzle_ready
--dont_report_missing_atoms
used only with --get_rnapuzzle_ready
--collapsed_view
--cv alias to collapsed_view
-v, --verbose tell me more what you're doing, please!
--replace_hetatm replace 'HETATM' with 'ATOM' [tested only with
--get_rnapuzzle_ready]
--inplace in place edit the file! [experimental, only for
get_rnapuzzle_ready, delete, get_ss, get_seq]
--mutate MUTATE mutate residues, e.g. A:1A+2A+3A+4A,B:1A to mutate the
first nucleotide of the A chain to Adenine etc and the
first nucleotide of the B chain
--edit EDIT edit 'A:6>B:200', 'A:2-7>B:2-7'
--replace-chain REPLACE_CHAIN
a file PDB name with one chain that will be used to
replace the chain in the original PDB file, the chain
id in this file has to be the same with the chain id
of the original chain
--delete DELETE delete the selected fragment, e.g. A:10-16, or for
more than one fragment --delete 'A:1-25+30-57'
--extract EXTRACT extract the selected fragment, e.g. A:10-16, or for
more than one fragment --extract 'A:1-25+30-57'
Tricks:
$ for i in *; do echo $i; rna_pdb_toolsx.py --delete A:48-52 $i > noloop/${i/.pdb/_noloop.pdb}; done
10_rp17c.out.14.pdb
10_rp17c.out.14_out.pdb
[..]
$ for i in *.pdb; do rna_pdb_toolsx.py --c $i > ${i/.pdb/_clx.pdb}; done
$ for i in *.pdb; do rna_pdb_toolsx.py --get_rnapuzzle_ready $i > ${i/.pdb/_rpr.pdb}; done
.. keep original structures in original and use rpr:
➜ bujnicki_server_ss for i in original/*.pdb; do rna_pdb_toolsx.py --get_rnapuzzle_ready $i > ${i/.pdb/_rpr.pdb}; done
➜ bujnicki_server_ss ls
17pz_withSS_all_thrs6.00A_clust01-000001_AA_rpr.pdb 17pz_withSS_all_thrs6.00A_clust06-000001_AA_rpr.pdb
17pz_withSS_all_thrs6.00A_clust02-000001_AA_rpr.pdb 17pz_withSS_all_thrs6.00A_clust07-000001_AA_rpr.pdb
17pz_withSS_all_thrs6.00A_clust03-000001_AA_rpr.pdb 17pz_withSS_all_thrs6.00A_clust08-000001_AA_rpr.pdb
17pz_withSS_all_thrs6.00A_clust04-000001_AA_rpr.pdb 17pz_withSS_all_thrs6.00A_clust09-000001_AA_rpr.pdb
17pz_withSS_all_thrs6.00A_clust05-000001_AA_rpr.pdb original
.. or to get SimRNAready structures:
$ for i in *pdb; do rna_pdb_toolsx.py --get_simrna_ready $i > ${i/.pdb/_srr.pdb}; done
See Utils for simple but still extremly powerful rna tools.
Read more http://rna-pdb-tools.readthedocs.io/en/latest/
Read the documentations at rna-pdb-tools.rtfd.io/en/latest/.
Read at https://rna-pdb-tools.readthedocs.io/en/latest/rna-puzzles.html
- https://www.rosettacommons.org/docs/latest/application_documentation/rna/RNA-tools
- http://blue11.bch.msu.edu/mmtsb/convpdb.pl
- https://github.com/haddocking/pdb-tools
- https://github.com/harmslab/pdbtools
- http://ginsberg.med.virginia.edu/Links/Phenix/pdbtools.htm
- .. and more!
Read at http://rna-pdb-tools.readthedocs.io/en/latest/install.html