SCATTR is a BIDS App that performs a tractography processing workflow to
identify connections between targetted structures of interest (extracted from
various atlases) in the brain, using some common neuroimaging tools like
prepdwi, Freesurfer, and Mrtrix3.
This is useful for:
- Analyzing structural connections of known brain circuits
- Examining specific connections of interest
- Comparison across different groups (e.g. controls vs patients)
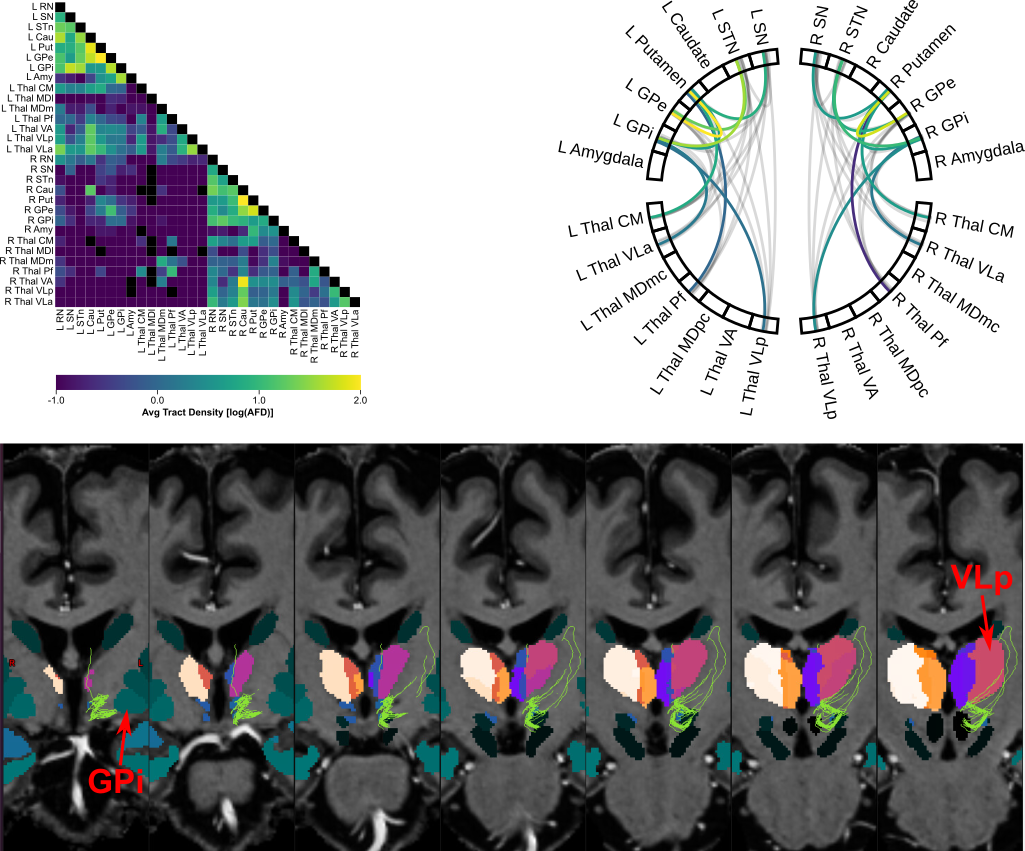
This workflow was used to process and analyze the data from
hcp_subcortical_repo
(see Kai et al., 2022).
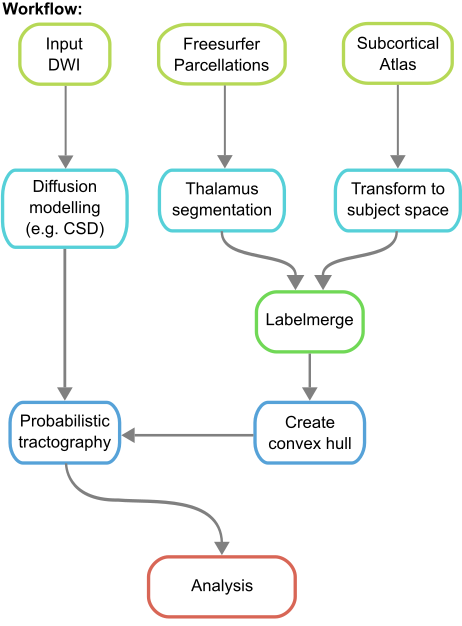
A brief summary of the workflow can be found below (see documentation for a detailed summary):
Note: The workflow assumes Freesurfer has already been run on the dataset, as well as diffusion preprocessing (e.g. distortion correction).
- Merge the segmentations of structures in a standard template space from various sources (if necessary) via labelmerge into a combined atlas, which is used downstream to identify targeted connections.
- Estimate and apply transformations from standard template space to subject-specific space.
- Further process the pre-processed diffusion data to enable tractography (e.g. compute response functions, fibre orientation distribution, normalization).
- Perform whole-brain tractography from computed files, applying filtering (e.g. SIFT2) to computed tractogram. Once filtered, connections between the merged segmentations (from step 1) are identified, generating a connectome map.
- Analysis can then be performed on the map to examine and explore the connectome of interest.
Full documentation: here
- Kai, J., Khan, A.R., Haast, R.A.M., Lau, J.C. (2022). Mapping the subcortical connectome using in vivo diffusion MRI: feasibility and reliability. Terra incognita: diving into the human subcortex, special issue of NeuroImage. doi: 10.1016/j.neuroimage.2022.119553.