papermill is a tool for parameterizing, executing, and analyzing Jupyter Notebooks.
Papermill lets you:
- parameterize notebooks
- execute and collect metrics across the notebooks
- summarize collections of notebooks
This opens up new opportunities for how notebooks can be used. For example:
- Perhaps you have a financial report that you wish to run with different values on the first or last day of a month or at the beginning or end of the year, using parameters makes this task easier.
- Do you want to run a notebook and depending on its results, choose a particular notebook to run next? You can now programmatically execute a workflow without having to copy and paste from notebook to notebook manually.
- Do you have plots and visualizations spread across 10 or more notebooks? Now you can choose which plots to programmatically display a summary collection in a notebook to share with others.
From the command line:
pip install papermill
For all optional io dependencies, you can specify individual bundles
like s3, or azure -- or use all
pip install papermill[all]
Other language bindings welcome if someone would like to maintain parallel implementations!
To parameterize your notebook designate a cell with the tag parameters.
Papermill looks for the parameters cell and treats this cell as defaults for the parameters passed in at execution time. Papermill will add a new cell tagged with injected-parameters with input parameters in order to overwrite the values in parameters. If no cell is tagged with parameters the injected cell will be inserted at the top of the notebook.
Additionally, if you rerun notebooks through papermill and it will reuse the injected-parameters cell from the prior run. In this case Papermill will replace the old injected-parameters cell with the new run's inputs.
The two ways to execute the notebook with parameters are: (1) through the Python API and (2) through the command line interface.
import papermill as pm
pm.execute_notebook(
'path/to/input.ipynb',
'path/to/output.ipynb',
parameters = dict(alpha=0.6, ratio=0.1)
)
Here's an example of a local notebook being executed and output to an Amazon S3 account:
$ papermill local/input.ipynb s3://bkt/output.ipynb -p alpha 0.6 -p l1_ratio 0.1
NOTE:
If you use multiple AWS accounts, and you have properly configured your AWS credentials, then you can specify which account to use by setting the AWS_PROFILE environment variable at the command-line. For example:
$ AWS_PROFILE=dev_account papermill local/input.ipynb s3://bkt/output.ipynb -p alpha 0.6 -p l1_ratio 0.1
In the above example, two parameters are set: alpha and l1_ratio using -p (--parameters also works). Parameter values that look like booleans or numbers will be interpreted as such. Here are the different ways users may set parameters:
$ papermill local/input.ipynb s3://bkt/output.ipynb -r version 1.0
Using -r or --parameters_raw, users can set parameters one by one. However, unlike -p, the parameter will remain a string, even if it may be interpreted as a number or boolean.
$ papermill local/input.ipynb s3://bkt/output.ipynb -f parameters.yaml
Using -f or --parameters_file, users can provide a YAML file from which parameter values should be read.
$ papermill local/input.ipynb s3://bkt/output.ipynb -y "
alpha: 0.6
l1_ratio: 0.1"
Using -y or --parameters_yaml, users can directly provide a YAML string containing parameter values.
$ papermill local/input.ipynb s3://bkt/output.ipynb -b YWxwaGE6IDAuNgpsMV9yYXRpbzogMC4xCg==
Using -b or --parameters_base64, users can provide a YAML string, base64-encoded, containing parameter values.
When using YAML to pass arguments, through -y, -b or -f, parameter values can be arrays or dictionaries:
$ papermill local/input.ipynb s3://bkt/output.ipynb -y "
x:
- 0.0
- 1.0
- 2.0
- 3.0
linear_function:
slope: 3.0
intercept: 1.0"
Papermill supports the following name handlers for input and output paths during execution:
-
Local file system:
local -
HTTP, HTTPS protocol:
http://, https:// -
Amazon Web Services: AWS S3
s3:// -
Azure: Azure DataLake Store, Azure Blob Store
adl://, abs:// -
Google Cloud: Google Cloud Storage
gs://
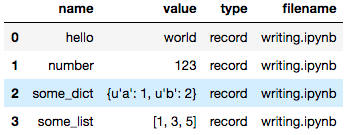
Users can save values to the notebook document to be consumed by other notebooks.
Recording values to be saved with the notebook.
"""notebook.ipynb"""
import papermill as pm
pm.record("hello", "world")
pm.record("number", 123)
pm.record("some_list", [1, 3, 5])
pm.record("some_dict", {"a": 1, "b": 2})
Users can recover those values as a Pandas dataframe via the
read_notebook function.
"""summary.ipynb"""
import papermill as pm
nb = pm.read_notebook('notebook.ipynb')
nb.dataframe
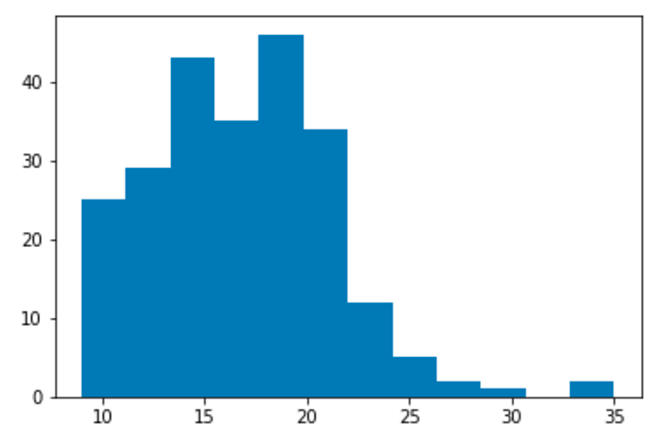
Display a matplotlib histogram with the key name matplotlib_hist.
"""notebook.ipynb"""
import papermill as pm
from ggplot import mpg
import matplotlib.pyplot as plt
# turn off interactive plotting to avoid double plotting
plt.ioff()
f = plt.figure()
plt.hist('cty', bins=12, data=mpg)
pm.display('matplotlib_hist', f)
Read in that above notebook and display the plot saved at
matplotlib_hist.
"""summary.ipynb"""
import papermill as pm
nb = pm.read_notebook('notebook.ipynb')
nb.display_output('matplotlib_hist')
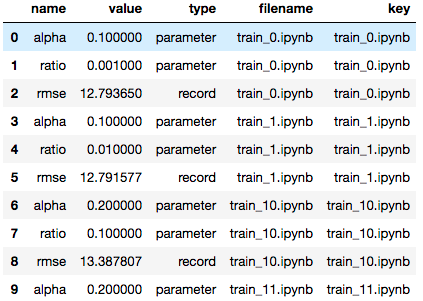
Papermill can read in a directory of notebooks and provides the
NotebookCollection interface for operating on them.
"""summary.ipynb"""
import papermill as pm
nbs = pm.read_notebooks('/path/to/results/')
# Show named plot from 'notebook1.ipynb'
# Accept a key or list of keys to plot in order.
nbs.display_output('train_1.ipynb', 'matplotlib_hist')
# Dataframe for all notebooks in collection
nbs.dataframe.head(10)
Read CONTRIBUTING.md for guidelines on how to setup a local development environment and make code changes back to Papermill.
For development guidelines look in the DEVELOPMENT_GUIDE.md file. This should inform you on how to make particular additions to the code base.
We host the Papermill documentation on ReadTheDocs.