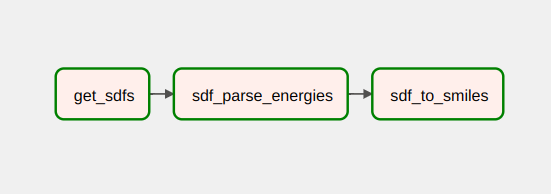
This repo was created to automate data conversion operations.
It helps to simply convert data from sdf format to smiles,
which can be used as an input to NLP models.
The pipeline was implemented with Apache Airflow [under the Docker].
The current pipeline consists of three steps:
- Merge all
.sdffiles into a single one - Get binding affinity from
.sdf - Convert compounds from
sdfto smiles, save tocsv(including binding energies)
To run the pipeline locally, you'll need to install:
- Docker Engine
- Docker Compose
First, you need to create an appropriate folder structure.
- create directories called
logsandpluginsin the project root - create
datafolder with the following structure: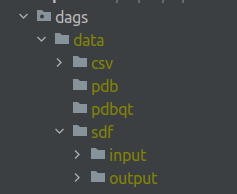
After that, place your compounds inside sdf/input folder. Then just open the terminal in the project root and type
docker-compose build
docker-compose up airflow-init
docker-compose upNote: you can change the default Airflow login and password inside
docker-compose.yamlif you want to.
For more detailed guide see: https://www.youtube.com/watch?v=aTaytcxy2Ck
And that's it, you can access http://localhost:8080/ with Airflow after these steps.
To run the pipeline, just hit the run DAG button :)
The pipeline will be extended soon. Planned features:
- Docking preparation tool (conversion to
pdbqt) - Data post-processing after SMILES generation