This project provides an implementation of Self-Organizing Map (SOM) in Go. It implements the two most well known SOM training algorithms: sequential and batch. The batch training is faster than the sequential as it can be parallelized, taking advantage of as many cores as your machine provides. However it can be less accurate as it merely provides a resonable approximation of SOM, but still acceptable. The sequential algorithm is performed as its name implies, sequentially. Because of its sequential nature it's slower than batch training, but more accurate. You can read more about SOM training algorithms here.
The goal of this project is to provide an API to build SOMs in Go. The project also implements various SOM quality measures which can help you validate the results of the training algorithm. In particular the project implements quantization and topographic error to measure both the projection and topography as well as topographic product which can help you make a decision about the size of the SOM grid.
Get the source code:
$ go get -u github.com/milosgajdos83/gosom
Run the tests:
$ make test
You can see the simplest example of SOM below:
func main() {
// make random data
d := []float64{5.1, 3.5, 1.4, 0.1,
4.9, 3.0, 1.4, 0.2,
4.7, 3.2, 1.3, 0.3,
4.6, 3.1, 1.5, 0.4,
5.0, 3.6, 1.4, 0.5}
data := mat64.NewDense(5, 4, d)
// SOM configuration
grid := &som.GridConfig{
Size: []int{2, 2},
Type: "planar",
UShape: "hexagon",
}
cb := &som.CbConfig{
Dim: 4,
InitFunc: som.RandInit,
}
mapCfg := &som.MapConfig{
Grid: grid,
Cb: cb,
}
// create new SOM
m, err := som.NewMap(mapCfg, data)
if err != nil {
fmt.Fprintf(os.Stderr, "\nERROR: %s\n", err)
os.Exit(1)
}
// training configuration
trainCfg := &som.TrainConfig{
Algorithm: "seq",
Radius: 500.0,
RDecay: "exp",
NeighbFn: som.Gaussian,
LRate: 0.5,
LDecay: "exp",
}
if err := m.Train(trainCfg, data, 300); err != nil {
fmt.Fprintf(os.Stderr, "\nERROR: %s\n", err)
os.Exit(1)
}
// check quantization error
qe, err := m.QuantError(data)
if err != nil {
fmt.Fprintf(os.Stderr, "\nERROR: %s\n", err)
os.Exit(1)
}
log.Printf("Quantization Error: %f\n", qe)
}If you build and run this program it will spit out quantization error. It's not that particularly exciting. You could generate a u-matrix, but since the data set is very simple, it would not be particularly interesting either. If you want to see more elaboarate and moreinteresting stuff you can do, check out the samples programs in examples directory.
SOMs are a very good tool to perform data clustering. Examples directory contains two more elaborate programs that illustrate the power of SOM clustering.
A classic schoolbook example of this is clustering of colors in arbitrary nosiy images. You can find a simple example program which does just this in the colors subdirectory of examples. When you build the program you can run it as follows:
$ make colors
$ ./_build/colors -umatrix umatrix.html -dims 40,40 -radius 500.0 -rdecay exp -lrate 0.5 -ldecay exp -ushape hexagon -iters 30000 -training seq -input ./examples/colors/testdata/colors.png -output som.png
[ gosom ] Loading data set ./examples/colors/testdata/colors.png
[ gosom ] Creating new SOM. Dimensions: [40 40], Grid Type: planar, Unit shape: hexagon
[ gosom ] Starting SOM training. Method: seq, iterations: 30000
[ gosom ] Training successfully completed. Duration: 3.843383347s
[ gosom ] Saving U-Matrix to umatrix.html
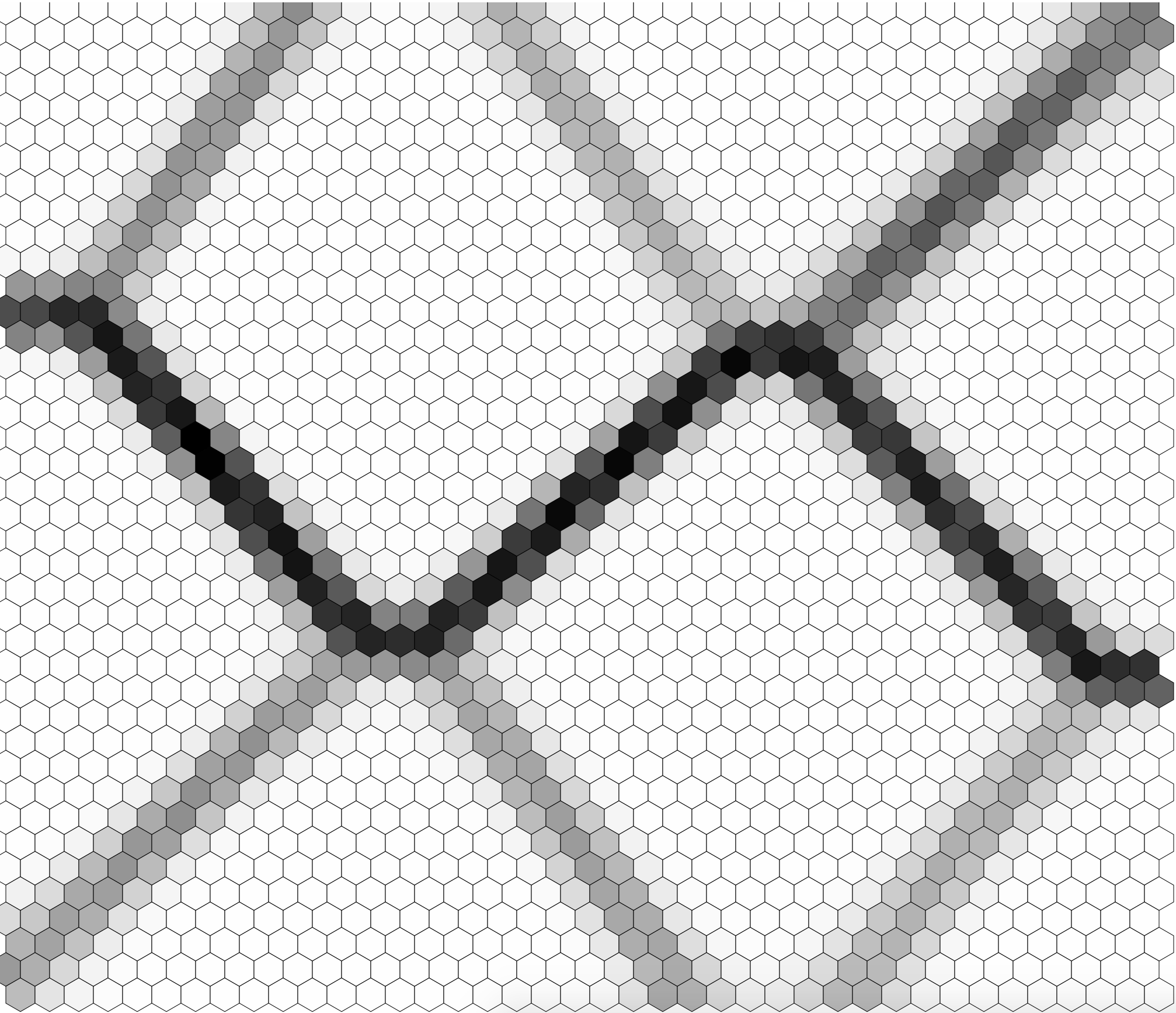
This program reads in a sample "noisy" image from the test data directory which looks like this:
It will spit out a new image som.png which looks like this:
Lastly, it will generate a u-matrix which looks like this:
Even more elaborate example can be found in fpcs directory. It is used to demostrate that both implemented algorithm behave as expected according to the following research. You can verify this yourself. First you have to build the fcps example program:
$ make fcps
The program provides various cli options:
$ ./_build/fcps -h
Examples of both batch and sequential training runs can be found below:
$ D=Target ./_build/fcps -umatrix umatrix_batch.html -dims 30,30 -radius 500.0 -rdecay exp -ushape rectangle -iters 100 -training batch -input examples/fcps/testdata/fcps/${D}.lrn -cls examples/fcps/testdata/fcps/${D}.cls
[ gosom ] Loading data set testdata/fcps/Target.lrn
[ gosom ] Creating new SOM. Dimensions: [30 30], Grid: planar, Unit shape: rectangle
[ gosom ] Starting SOM training. Method: batch, iterations: 100
[ gosom ] Training successfully completed. Duration: 1.923548243s
[ gosom ] Saving U-Matrix to umatrix_batch.html
[ gosom ] Quantization Error: 0.023212
[ gosom ] Topographic Product: +Inf
[ gosom ] Topographic Error: 0.015584
$ D=Target ./_build/fcps -umatrix umatrix_seq.html -dims 30,30 -radius 500.0 -rdecay exp -lrate 0.5 -ldecay exp -ushape hexagon -iters 30000 -training seq -input examples/fcps/testdata/fcps/${D}.lrn -cls examples/fcps/testdata/fcps/${D}.cls
[ gosom ] Loading data set testdata/fcps/Target.lrn
[ gosom ] Creating new SOM. Dimensions: [30 30], Grid: planar, Unit shape: hexagon
[ gosom ] Starting SOM training. Method: seq, iterations: 30000
[ gosom ] Training successfully completed. Duration: 2.261068582s
[ gosom ] Saving U-Matrix to umatrix_seq.html
[ gosom ] Quantization Error: 0.064704
[ gosom ] Topographic Product: 0.010281
[ gosom ] Topographic Error: 0.014286
Both of the above mentioned runs generate a simple umatrix that displays the clustered data in svg format. You can now inspect the files to cmpare the both algorithms.
Test data present in fcps subdirectory of testdata come from Philipps University of Marburg:
Ultsch, A.: Clustering with SOM: U*C, In Proc. Workshop on Self-Organizing Maps, Paris, France, (2005) , pp. 75-82