This repository contains the end-to-end pipeline for skull-stripping of T1 Weighted MRI images and Canonical ICA of resting state fMRI (rs-fMRI) to achieve the Default Mode Network (DMN) of the brain.
Follow the below steps to setup the machine to run this project.
Download the skull-stripped images dataset from the NFBS repository and extract the tar.gz file to get the dataset under NFBS_Dataset directory.
Run the following command to install and create a virtual environment for the project. Run all these commands from inside the repository folder.
Clone to repository
git clone https://github.com/aksh-ai/skull-stripping-and-ica.git
Install virtual environment library
pip install --user virtualenv
Create a virtual environment named brain
python -m venv brain
To activate the vrtual environment
.\env\Scripts\activate
To deactivate the virtual environment
deactivate
Install the required libraries using the following command
pip install --upgrade -r requirements.txt
There are several scripts available in the repository that can be used to train and run inference for skull-stripping and canonical ICA. The parameters given in examples for the scripts were the parameters used for training, evaluation, and testing of the models to achieve the required results.
The following scripts can be used to train, and run inference for skull-stripping
The data can be prepared in the required format using the prepare_data.py script. It takes the following arguments
python prepare_data.py -r ROOT_DIR -o CSV_FILE_NAME -d OUTPUT_DIR
Example:
python prepare_data.py -r 'NFBS_Dataset' -o 'NFBS_Dataset_meta.csv' -d 'data'
Refer data_exp.ipynb to run the same but in an interactive manner.
The train.py script can be used to train and evaluate the skull-stripping Residual UNET 3D model
python train.py \
-i IMAGES_DIR \
-l MASK_DIR \
-m MODEL_DIR \
-ic INPUT_CHANNELS \
-oc OUTPUT_CHANNELS \
-d DEVICE \
-mn MODEL_NAME \
-p PATCH_SIZE \
-b BATCH_SIZE \
-hl HISTOGRAM_LANDMARKS \
-e EPOCHS \
-ls LOSS \
-opt OPTIMIZER \
-lr LEARNING_RATE \
-s SCHEDULER \
-sk OPTIONAL_SKIP \
-t TEST_SIZE \
-tb TENSORBOARD \
-c CHECKPOINT \
-log TENSOBOARD_LOG_DIR \
-v VERBOSE \
-mp MODEL_CHECKPOINT_PATH
Example:
python train.py -i 'data/images' -l 'data/targets' -m 'models' -ic 1 -oc 1 -d 'cuda' \
-mn 'res_unet_3d' -p 64 -b 6 -hl 'NFBS_histogram_landmarks.npy' -e 6 -ls 'MSE' \
-opt 'Adam' -s True -sk False -t 0.2 -tb True -c True -log 'ss_trianing_logs' -v 400
Refer skull_stripping_training_MSE.ipynb notebook which was run on Google Colab for the training and evaluation of the model (without optional skip)
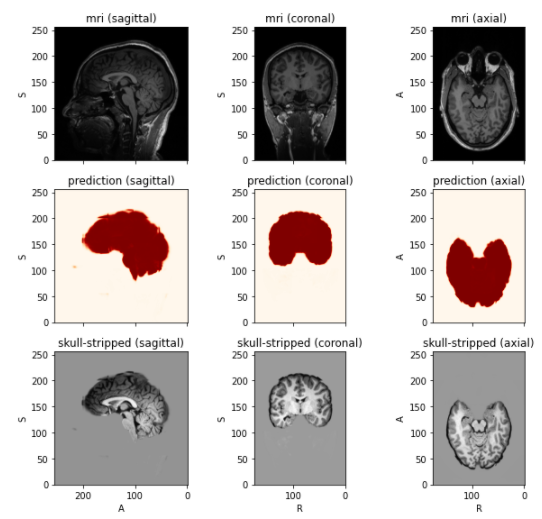
The inference.py script can be used to run inference on a new T1 Weighted MRI Image which outputs and saves the skull-stripped image. The model used for this is residual_unet3d_MSE_2.pth present under the models directory
python inference.py -i INPUT_IMAGE -o OUTPUT_IMAGE_NAME -d DEVICE -m MODEL_PATH -p PATCH_SIZE -l OVERLAP_SIZE -b BATCH_SIZE -v VISUALIZE -s OPTIONAL_SKIP
Example:
python inference.py -i 'T1Img\sub-02\anat_img.nii.gz' -o 'sub-02-anat-img-skull-stripped.nii.gz' -d 'cuda' -m 'models\residual_unet3d_MSE_2.pth' -p 64 -l 16 -b 1 -v True -s False
Refer inference.ipynb notebook for the inference running the inference on interactive python notebooks
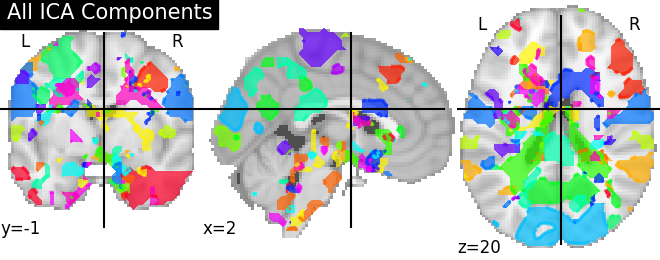
The ica.py script can be used to perform Canonical ICA for resting state MRI images to decompose the rs-fMRI scans into different componenets/networks of the brain.
python ica.py -i INPUT_PATH -o OUTPUT_PATH -n NUM_COMPONENTS -v VERBOSE -vis VISUALIZE -m MEMORY_LEVEL
Example:
python ica.py -i 'Filtered_4DVolume.nii' -o 'ica_components.nii' -n 20 -v 10 -vis True -m 2
Refer ica.ipynb for the exploratory experiemntation of the ICA components of the rs-fMRI image to achieve Default Mode Network (DMN)
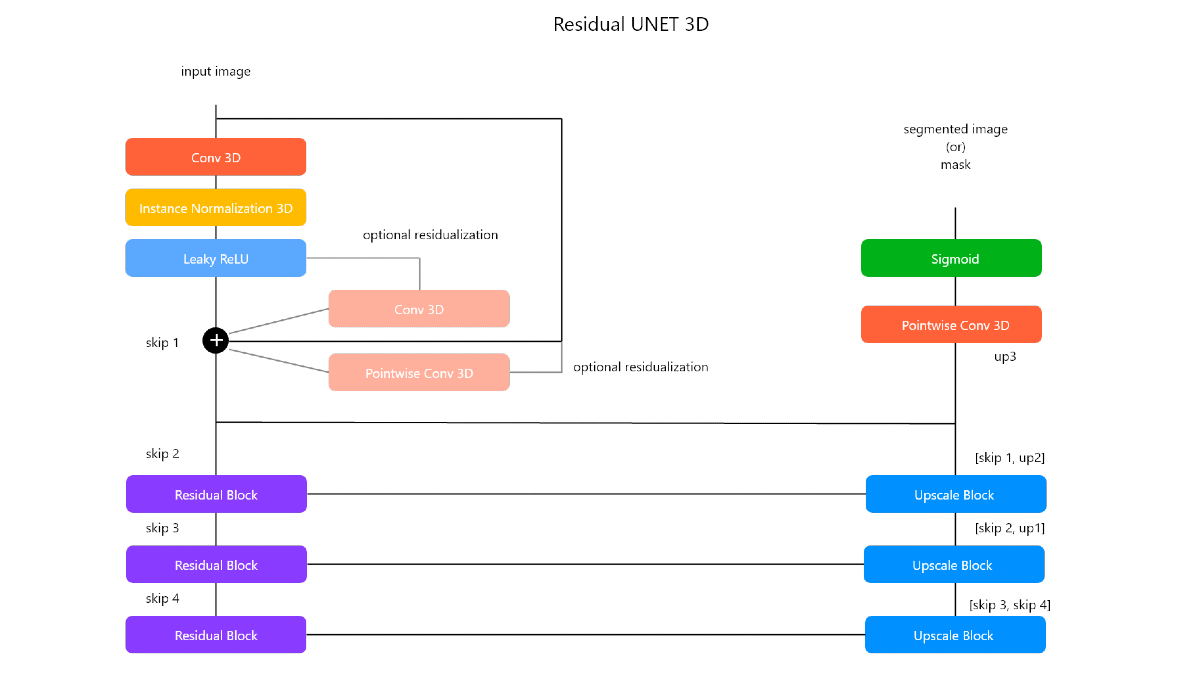
Residual UNET 3D is a custom image segmentation architecture inspired from Residual Networks and UNET for 3D images. Below is the architecture diagram of the model.
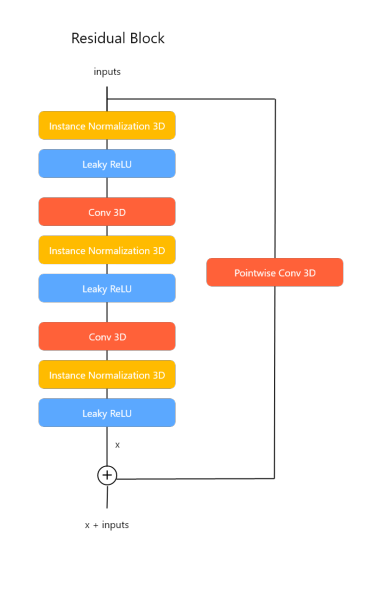
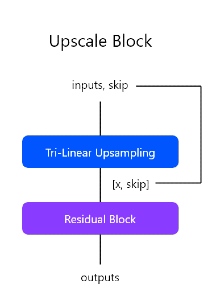
The architecture basically consists of two important blocks:
- Residual Block
- Upscaling Block
The lib library contains the following modules which is reusable and enables to implement the required fucntionalities for the skull-stripping in just a few lines of code.
- data - Requires the functions to load the NFBS datasets as whoel iamges and sub-volumes
- layers - Contains the neural network layers required for implementing the neural network architectures
- losses - Contains a collection of loss functions that can be used for image segmentation, generative adversarial networks, and variational autoencoders
- models - Contains the Residual UNET 3D image segmentation architecture
- runners - Contains the functions for training, evaluation, and inference
- utils - Contains functions related data preparation, image visualization, preporcessing, augmentations, and segmetation operations
The notebooks contain the data exploration, data pipeline description, training, evaluation, and testing of the models
- data_exp.ipynb - This notebook contains the meta data csv preparation of the NFBS dataset for data preparation
- data_exploration_playground.ipynb - This notebook contains the data exploration of MRI images by visualizing them
- data_pipeline_testing.ipynb - This notebook contains the different data pipeline's (whole and sub-volumes) usages using the
datamodule fromlib - skull_stripping_training_MSE.ipynb - This notebook contains the training, evaluation, and performance on the test set using the Residual UNET 3D for skull-stripping without the optional skip
- skull_stripping_training_MSE_3.ipynb - This notebook contains the training, evaluation, and performance on the test set using the Residual UNET 3D for skull-stripping with the optional skip
- inference.ipynb - This notebook contains the inference pipeline in notebook format using the
inference.pyscript - ica.ipynb - This nontebook contains the epxloratory expeimentation of ICA to achieve DMN
The skull-stripped images for the T1 Weighted MRI test set and the Canonical ICA components image (probability atlas) for the rs-fMRI image is available at this google drive link
- Skull-stripping on test image
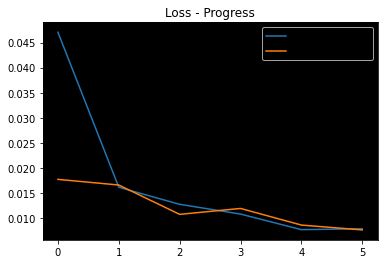
- Skull-stripping MSE Loss progress
Without optional skip:
Blue - Training Loss
Orange - Validation Loss
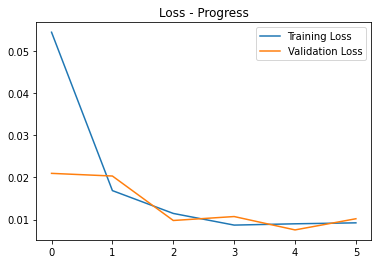
With optional skip:
Blue - Training Loss
Orange - Validation Loss
- Metrics
| Model Name | Optional Skip | MSE Loss | Dice Score | IoU Score |
|---|---|---|---|---|
| models\residual_unet3d_MSE_2.pth | False | 0.008068 | 91.76 % | 87.53 % |
| models\residual_unet3d_MSE_3.pth | True | 0.010058 | 91.13 % | 86.11 % |
- Canonical ICA components image
- Deep Residual Learning for Image Recognition
- U-Net: Convolutional Networks for Biomedical Image Segmentation
- DoubleU-Net: A Deep Convolutional Neural Network for Medical Image Segmentation
- Vox2Vox: 3D-GAN for Brain Tumour Segmentation
- An automated method for identifying an independent component analysis-based language-related resting-state network in brain tumor subjects for surgical planning
- Age and Alzheimer’s pathology disrupt default mode network functioning via alterations in white matter microstructure but not hyperintensities