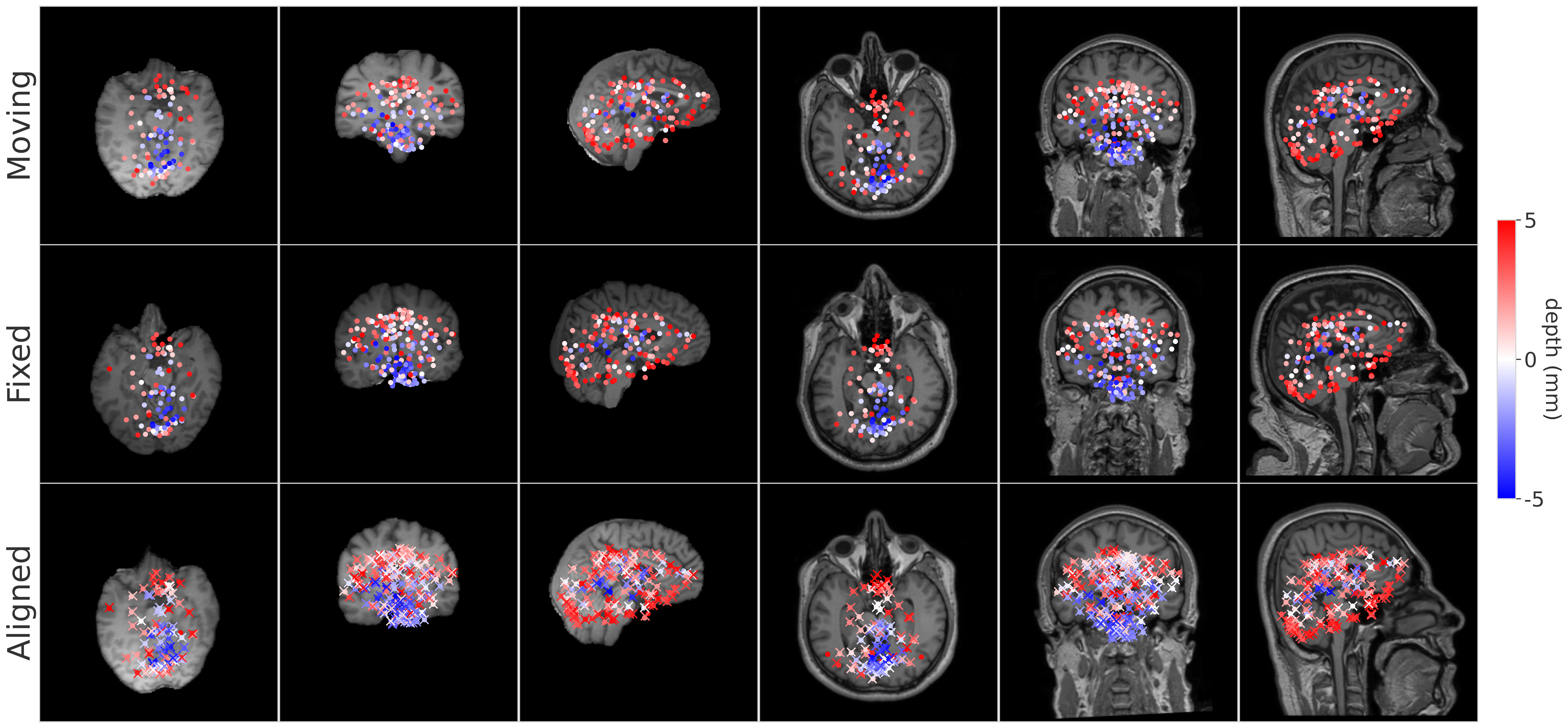
BrainMorph is a foundation model for brain MRI registration. It is a deep learning-based model trained on over 100,000 brain MR images at full resolution (256x256x256). The model is robust to normal and diseased brains, a variety of MRI modalities, and skullstripped and non-skullstripped images. It supports unimodal/multimodal pairwise and groupwise registration using rigid, affine, or nonlinear transformations.
BrainMorph is built on top of the KeyMorph framework, a deep learning-based image registration framework that relies on automatically extracting corresponding keypoints.
Check out the colab tutorial to get started!
- [May 2024] The preprint for BrainMorph is available on arXiv!
- [May 2024] Released full set of BrainMorph models on Box. Usage instructions under "Registering brain volumes" (paper to come!).
git clone https://github.com/alanqrwang/brainmorph.git
cd brainmorph
pip install -e .The brainmorph package depends on the following requirements:
- keymorph>=1.0.0
- numpy>=1.19.1
- ogb>=1.2.6
- outdated>=0.2.0
- pandas>=1.1.0
- pytz>=2020.4
- torch>=1.7.0
- torchvision>=0.8.2
- scikit-learn>=0.20.0
- scipy>=1.5.4
- torchio>=0.19.6
Running pip install -e . will automatically check for and install all of these requirements.
The --download flag in the provided script will automatically download the corresponding model and place is in the folder specified by --weights_dir (see below commands).
Otherwise, you can find all BrainMorph trained weights here and manually place them in the folder specified by --weights_dir.
To get started, check out the colab tutorial!
The script will automatically min-max normalize the images and resample to 1mm isotropic resolution.
--num_keypoints and --variant will determine which model will be used to perform the registration.
--num_keypoints can be set to 128, 256, 512 and --variant can be set to S, M, L (corresponding to model size).
To register a single pair of volumes:
python scripts/register.py \
--num_keypoints 256 \
--variant S \
--weights_dir ./weights/ \
--moving ./example_data/img_m/IXI_000001_0000.nii.gz \
--fixed ./example_data/img_m/IXI_000002_0000.nii.gz \
--moving_seg ./example_data/seg_m/IXI_000001_0000.nii.gz \
--fixed_seg ./example_data/seg_m/IXI_000002_0000.nii.gz \
--list_of_aligns rigid affine tps_1 \
--list_of_metrics mse harddice \
--save_eval_to_disk \
--save_dir ./register_output/ \
--visualize \
--download
Description of other important flags:
--movingand--fixedare paths to moving and fixed images.--moving_segand--fixed_segare paths to moving and fixed segmentation maps. These are optional, but are required if you want the script to report Dice scores or surface distances.--list_of_alignsspecifies the types of alignment to perform. Options arerigid,affineandtps_<lambda>(TPS with hyperparameter value equal to lambda). lambda=0 corresponds to exact keypoint alignment. lambda=10 is very similar to affine.--list_of_metricsspecifies the metrics to report. Options aremse,harddice,softdice,hausd,jdstd,jdlessthan0. To compute Dice scores and surface distances,--moving_segand--fixed_segmust be provided.--save_eval_to_disksaves all outputs to disk.--save_dirspecifies the folder where outputs will be saved. The default location is./register_output/.--visualizeplots a matplotlib figure of moving, fixed, and registered images overlaid with corresponding points.--downloaddownloads the corresponding model weights automatically if not present in--weights_dir.
You can also replace filenames with directories to register all pairs of images in the directories. Note that the script expects corresponding image and segmentation pairs to have the same filename.
python scripts/register.py \
--num_keypoints 256 \
--variant S \
--weights_dir ./weights/ \
--moving ./example_data/img_m/ \
--fixed ./example_data/img_m/ \
--moving_seg ./example_data/seg_m/ \
--fixed_seg ./example_data/seg_m/ \
--list_of_aligns rigid affine tps_1 \
--list_of_metrics mse harddice \
--save_eval_to_disk \
--save_dir ./register_output/ \
--visualize \
--downloadTo register a group of volumes, put the volumes in ./example_data/img_m. If segmentations are available, put them in ./example_data/seg_m. Then run:
python scripts/register.py \
--groupwise \
--num_keypoints 256 \
--variant S \
--weights_dir ./weights/ \
--moving ./example_data/ \
--fixed ./example_data/ \
--moving_seg ./example_data/ \
--fixed_seg ./example_data/ \
--list_of_aligns rigid affine tps_1 \
--list_of_metrics mse harddice \
--save_eval_to_disk \
--save_dir ./register_output/ \
--visualize \
--downloadHere's a pseudo-code version of the registration pipeline that BrainMorph uses.:
def forward(img_f, img_m, seg_f, seg_m, network, optimizer, kp_aligner):
'''Forward pass for one mini-batch step.
Variables with (_f, _m, _a) denotes (fixed, moving, aligned).
Args:
img_f, img_m: Fixed and moving intensity image (bs, 1, l, w, h)
seg_f, seg_m: Fixed and moving one-hot segmentation map (bs, num_classes, l, w, h)
network: Keypoint extractor network
kp_aligner: Rigid, affine or TPS keypoint alignment module
'''
optimizer.zero_grad()
# Extract keypoints
points_f = network(img_f)
points_m = network(img_m)
# Align via keypoints
grid = kp_aligner.grid_from_points(points_m, points_f, img_f.shape, lmbda=lmbda)
img_a, seg_a = utils.align_moving_img(grid, img_m, seg_m)
# Compute losses
mse = MSELoss()(img_f, img_a)
soft_dice = DiceLoss()(seg_a, seg_f)
if unsupervised:
loss = mse
else:
loss = soft_dice
# Backward pass
loss.backward()
optimizer.step()The network variable is a CNN with center-of-mass layer which extracts keypoints from the input images.
The kp_aligner variable is a keypoint alignment module. It has a function grid_from_points() which returns a flow-field grid encoding the transformation to perform on the moving image. The transformation can either be rigid, affine, or nonlinear (TPS).
Use scripts/run.py with --run_mode train to train BrainMorph.
If you want to train with your own data, we recommend starting with the more minimal keymorph repository.
This repository is being actively maintained. Feel free to open an issue for any problems or questions.
If this code is useful to you, please consider citing the BrainMorph paper.
Alan Q. Wang, et al. "BrainMorph: A Foundational Keypoint Model for Robust and Flexible Brain MRI Registration."