Official Repo of the paper DeepMiCa: automatic segmentation and classification of breast MIcroCAlcifications from mammograms - Alessia Gerbasi et al.
Breast microcalcifications may be the very early and only first detectable sign of breast cancer. However, they are by definition small calcium deposits with less than 1 mm in diameter, and their early detection and classification is still a very challenging task, often requiring an invasive biopsy.
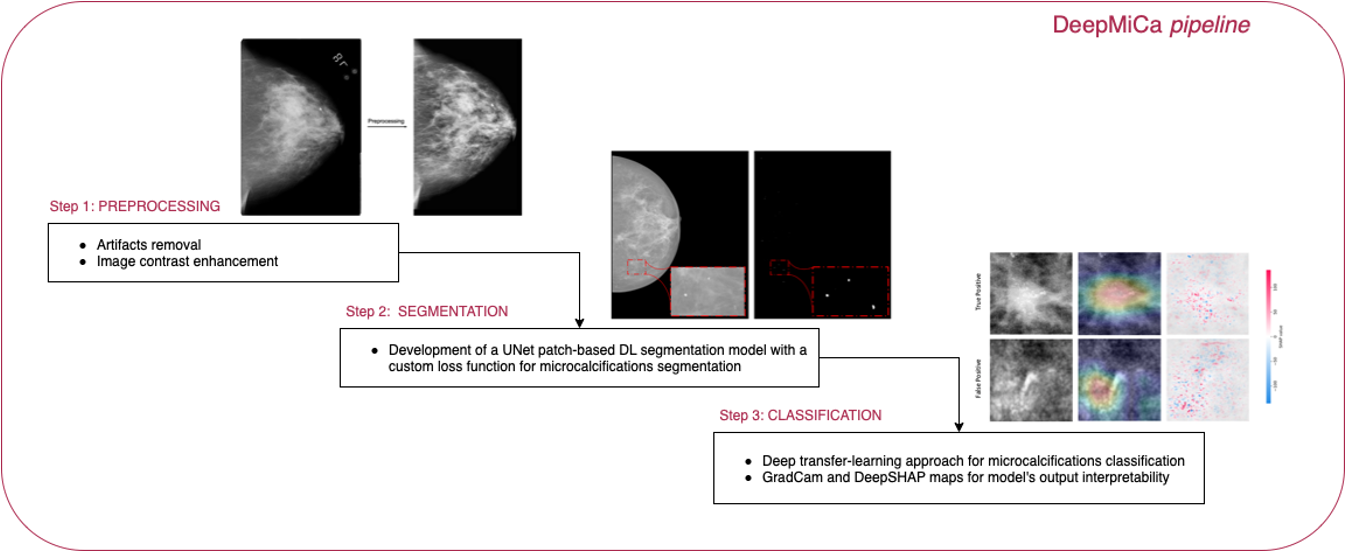
DeepMiCa can help to automate the whole process with a customisable accurate and visually explainable deep learning pipeline designed to support the clinicians during breast screening. The proposed pipeline pre-processes, segments and classifies microcalcifications from mammograms. The main steps are schematically represented in the following figure. Please refer to the paper for more details.
python > 3.7.
All the required packages are listed in the requirements.txt file.
The datasets used in this work are publicly available at the Curated Breast Imaging Subset of DDSM (CBIS-DDSM) and the INbreast. For a full description of the datasets, please refer to the paper and the original sources.
This is the tree structure of the folders you should have to directly use our code:
├── Step1_Preprocessing
│ └── preprocessing.py
│
├── Step2_Segmentation
│ ├── _01_SplitData
│ │ └── split.py
│ ├── _02_Patches
│ │ ├── create_patches.py
│ │ └── reduce_patches.py
│ ├── _03_Train
│ │ ├── LoadData.py
│ │ ├── Losses.py
│ │ ├── main.py
│ │ ├── SegmentationDataset.py
│ │ ├── train.py
│ │ └── UNet.py
│ ├── _04_Test
│ │ └── testing.py
│ └── Checkpoints
│
├── Step3_Classification
│ ├── _01_CutROI
│ │ └── cut_ROI.py
│ ├── _02_SplitData
│ │ └── split.py
│ ├── _03_FeatureExtraction
│ │ ├── CustomDataset.py
│ │ ├── LoadData.py
│ │ ├── main.py
│ │ ├── Resnet18.py
│ │ ├── train.py
│ │ └── Vgg16.py
│ ├── _04_FineTuning
│ │ └── fineTuning.py
│ └── _05_Test
│ └── testing.py
│
└── Datasets
├── INbreast
│ ├── AllPng
│ │ ├── 20586908.png
│ │ ├── 20586934.png
│ │ ├── ...
│ ├── Masks
│ │ ├── 20586908.png
│ │ ├── 20586934.png
│ │ ├── ...
│ └── csv
│ └── INbreast_table_noClusters.csv
└── CBIS_DDSM
├── AllPng
│ ├── 00005_RIGHT_CC.png
│ ├── 00007_LEFT_CC.png
│ ├── ...
├── Masks
│ ├── 00005_RIGHT_CC_1.png
│ ├── 00007_LEFT_CC_1.png
│ ├── ...
└── csv
└── CBIS_DDSM.csv
Our pipeline is designed to be as flexible as possible therefore, we provide a set of progressively numbered folders containing the scripts for each step. According to your needs, you can choose to run all the steps or jump to the one you are interested in. If your input dataset is different, or you did not structure the data folders as we did, you can easily change the paths in the scripts.
-
Step1_Preprocessing
preprocessing.py: this step is designed to preprocess the images from both datasets before the segmentation step in order to remove artifacts and enhance the contrast.
-
Step2_Segmentation
- _01_SplitData
split.py: splits the images from INbreast dataset into training, validation and test sets.
- _02_Patches
create_patches.py: create 256x256 pixels patches from images of both datasets.reduce_patches.py: you can run this script to remove the completely black patches from the training set.
- _03_Train
main.py: this is the main script to train the model that recalls all the other files in the folder. You can visualize the learning curves with wandb while training. The best weights will be saved in a folder calledCheckpoints.
- _04_Test
test.py: you can call this script to test the model on the test set.
- _01_SplitData
-
Step3_Classification
- _01_CutROI
cut_roi.py: in this script we cut the minimum bounding box including the segmented microcalcification from CBIS-DDSM dataset.
- _02_SplitData
split.py: splits the ROIs to be classified into training, validation and test sets.
- _03_FeatureExtraction
main.py: this is the main script to train the model that recalls all the other files in the folder. The best weights will be saved in a folder calledCheckpoints. You can visualize the learning curves with wandb while training. For hyperparameters tuning with RayTune, please refer to the RayTune documentation.
- _04_FineTuning
finetuning.py: this is the main script to fine-tune the model that recalls all the other files in the folder. The best weights will be saved in a folder calledCheckpoints. You can visualize the learning curves with wandb while training.
- _01_CutROI
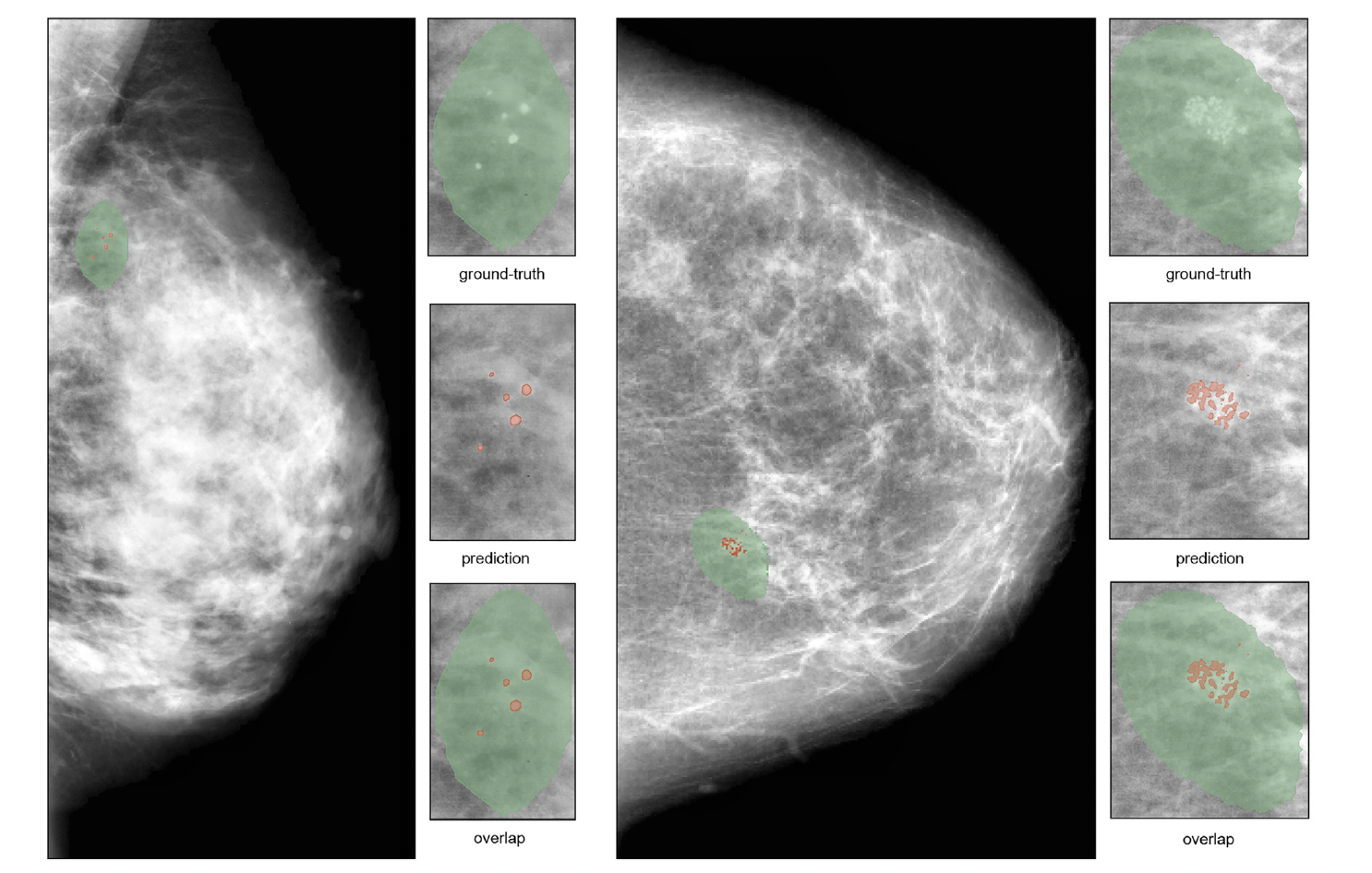
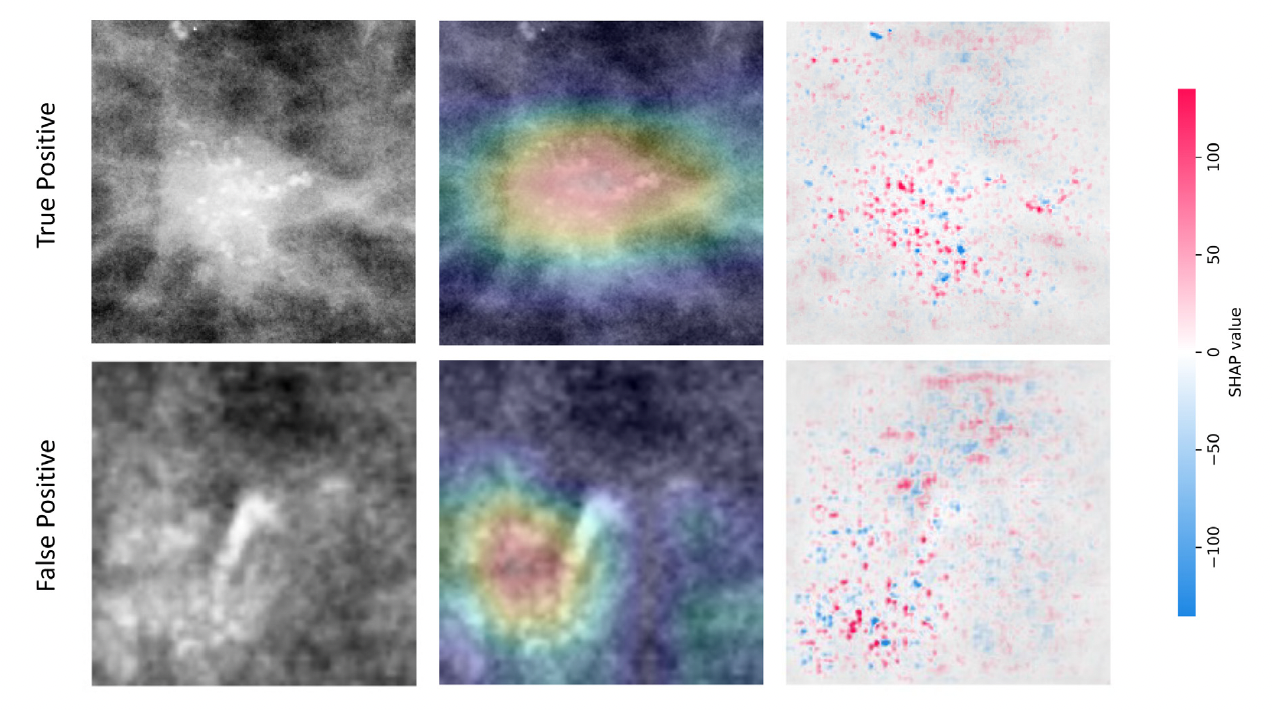
For a rapid visual assessment of the results we show here (1) A comparison between original CBIS-DDSM ground-truth masks and our segmentation results and (2) two examples of classification results and corresponding XAI maps. Please refer to the paper for a full description and discussion of the results.
If you want to directly test our models, you can request the models weights here and put them in the Checkpoints folder.
Then you can run the following scripts:
python3 Step2_Segmentation/_04_Test/testing.pyto test the segmentation model on your test set.python3 Step3_Classification/_05_Test/testing.pyto test the classification model on your test set.
If you find this code useful for your research, please cite our paper:
@article{gerbasi2023deepmica,
title={DeepMiCa: Automatic segmentation and classification of breast MIcroCAlcifications from mammograms},
author={Gerbasi, Alessia and Clementi, Greta and Corsi, Fabio and Albasini, Sara and Malovini, Alberto and Quaglini, Silvana and Bellazzi, Riccardo},
journal={Computer Methods and Programs in Biomedicine},
volume={235},
pages={107483},
year={2023},
publisher={Elsevier}
}