papaja is an award-winning R package that facilitates creating computationally reproducible, submission-ready manuscripts which conform to the American Psychological Association (APA) manuscript guidelines (6th Edition). papaja provides
- an R Markdown template that can be used with (or without) RStudio to create PDF documents (using the apa6 LaTeX class) or Word documents (using a .docx-reference file).
- Functions to typeset the results from statistical analyses,
- functions to create tables, and
- functions to create figures in accordance with APA guidelines.
For a comprehensive introduction to papaja, see the current draft of the manual. If you have a specific question that is not answered in the manual, feel free to ask a question on Stack Overflow using the papaja tag. If you believe you have found a bug or would like to request a new feature, open an issue on Github and provide a minimal complete verifiable example.
Take a look at the source file of the package vignette and the resulting PDF. The vignette also contains some basic instructions.
To use papaja you need either a recent version of RStudio or pandoc. If you want to create PDF- in addition to DOCX-documents you additionally need a TeX distribution. We recommend you use TinyTex, which can be installed from within R:
if(!requireNamespace("tinytex", quietly = TRUE)) install.packages("tinytex")
tinytex::install_tinytex()You may also consider MikTeX for Windows, MacTeX for Mac, or TeX Live for Linux. Please refer to the papaja manual for detailed installation instructions.
papaja is available on CRAN but you can also install it from the GitHub repository:
# Install latest CRAN release
install.packages("papaja")
# Install remotes package if necessary
if(!requireNamespace("remotes", quietly = TRUE)) install.packages("remotes")
# Install the stable development version from GitHub
remotes::install_github("crsh/papaja")
# Install the latest development snapshot from GitHub
remotes::install_github("crsh/papaja@devel")Once papaja is installed, you can select the APA template when creating a new R Markdown file through the RStudio menus.
To add citations, specify your bibliography-file in the YAML front
matter of the document (bibliography: my.bib) and start citing (for
details, see pandoc manual on the citeproc
extension. You may
also be interested in citr, an R
Studio addin to swiftly insert Markdown citations and R Studio’s visual
editor, which also
enables swiftly inserting
citations.
The functions apa_print() and apa_table() facilitate reporting
results of your analyses. When you pass the an output object of a
supported class, such as an htest- or lm-object, to apa_print(),
it will return a list of character strings that you can use to report
the results of your analysis.
my_lm <- lm(
Sepal.Width ~ Sepal.Length + Petal.Width + Petal.Length
, data = iris
)
apa_lm <- apa_print(my_lm)
apa_lm$full_result$Sepal_Length## [1] "$b = 0.61$, 95\\% CI $[0.48, 0.73]$, $t(146) = 9.77$, $p < .001$"
papaja currently provides methods for the following object classes:
| A-B | D-L | L-S | S-Z |
|---|---|---|---|
| afex_aov | default | lsmobj | summary.aovlist |
| anova | emmGrid | manova | summary.glht |
| anova.lme | glht | merMod | summary.glm |
| Anova.mlm | glm | mixed | summary.lm |
| aov | htest | papaja_wsci | summary.manova |
| aovlist | list | summary_emm | summary.ref.grid |
| BFBayesFactor | lm | summary.Anova.mlm | |
| BFBayesFactorTop | lme | summary.aov |
apa_table() may be used to produce publication-ready tables in an R
Markdown document. For instance, you might want to report some condition
means (with standard errors).
library("dplyr")
npk |>
group_by(N, P) |>
summarise(mean = mean(yield), se = sd(yield) / sqrt(length(yield)), .groups = "drop") |>
label_variables(N = "Nitrogen", P = "Phosphate", mean = "*M*", se = "*SE*") |>
apa_table(caption = "Mean pea yield (with standard errors)")Table 1. Mean pea yield (with standard errors)
| Nitrogen | Phosphate | M | SE |
|---|---|---|---|
| 0 | 0 | 51.72 | 1.88 |
| 0 | 1 | 52.42 | 2.65 |
| 1 | 0 | 59.22 | 2.66 |
| 1 | 1 | 56.15 | 2.08 |
This is a fairly simple example, but apa_table() may be used to
generate more complex tables.
apa_table(), of course, plays nicely with the output from
apa_print(). Thus, it is possible to conveniently report complete
regression tables, ANOVA tables, or the output from mixed-effects
models.
lm(Sepal.Width ~ Sepal.Length + Petal.Width + Petal.Length, data = iris) |>
apa_print() |>
apa_table(caption = "Iris regression table.")Table 2. Iris regression table.
| Predictor | b | 95% CI | t | df | p |
|---|---|---|---|---|---|
| Intercept | 1.04 | [0.51, 1.58] | 3.85 | 146 | < .001 |
| Sepal Length | 0.61 | [0.48, 0.73] | 9.77 | 146 | < .001 |
| Petal Width | 0.56 | [0.32, 0.80] | 4.55 | 146 | < .001 |
| Petal Length | -0.59 | [-0.71, -0.46] | -9.43 | 146 | < .001 |
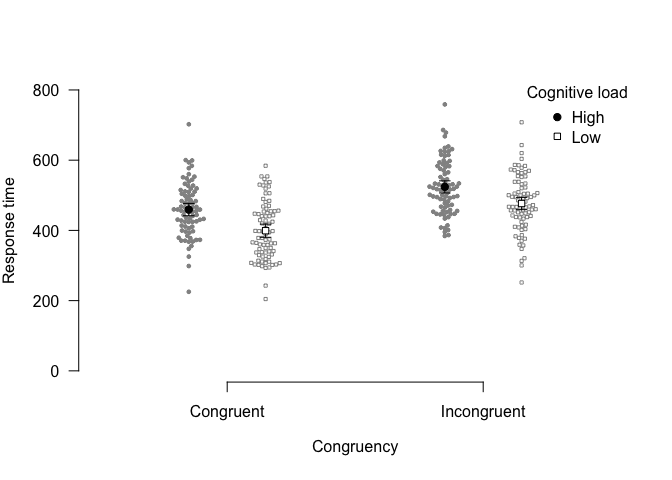
papaja further provides functions to create publication-ready plots.
For example, you can use apa_barplot(), apa_lineplot(), and
apa_beeplot() (or the general function apa_factorial_plot()) to
visualize the results of factorial study designs:
apa_beeplot(
data = stroop_data
, dv = "response_time"
, id = "id"
, factors = c("congruency", "load")
, ylim = c(0, 800)
, dispersion = wsci # within-subjects confidence intervals
, conf.level = .99
, las = 1
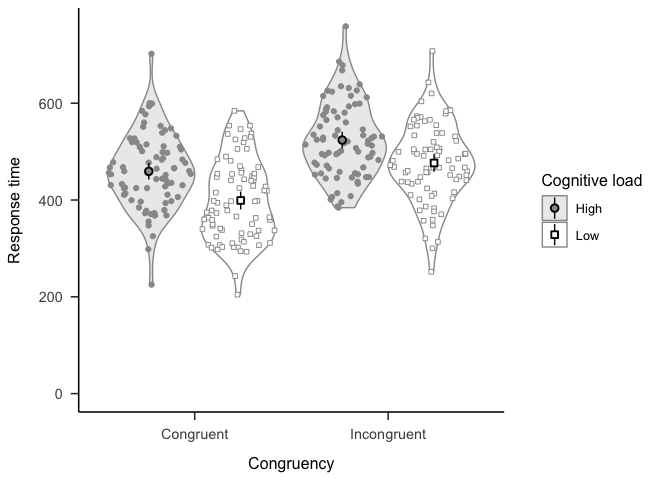
)If you prefer ggplot2, try theme_apa().
library("ggplot2")
library("ggforce")
p <- ggplot(
stroop_data
, aes(x = congruency, y = response_time, shape = load, fill = load)
) +
geom_violin(alpha = 0.2, color = grey(0.6)) +
geom_sina(color = grey(0.6)) +
stat_summary(position = position_dodge2(0.95), fun.data = mean_cl_normal) +
lims(y = c(0, max(stroop_data$response_time))) +
scale_shape_manual(values = c(21, 22)) +
scale_fill_grey(start = 0.6, end = 1) +
labs(
x = "Congruency"
, y = "Response time"
, shape = "Cognitive load"
, fill = "Cognitive load"
)
p + theme_apa()Don’t use RStudio? No problem. Use the rmarkdown::render function to
create articles:
# Create new R Markdown file
rmarkdown::draft(
"mymanuscript.Rmd"
, "apa6"
, package = "papaja"
, create_dir = FALSE
, edit = FALSE
)
# Render manuscript
rmarkdown::render("mymanuscript.Rmd")For a comprehensive introduction to papaja, check out the current draft of the papaja manual. If you have a specific question that is not answered in the manual, feel free to ask a question on Stack Overflow using the papaja tag. If you believe you have found a bug or you want to request a new feature, open an issue on Github and provide a minimal complete verifiable example.
Please cite papaja if you use it.
Aust, F. & Barth, M. (2022). papaja: Prepare reproducible APA journal articles with R Markdown. R package version 0.1.0.9999. Retrieved from https://github.com/crsh/papaja
For convenience, you can use
cite_r()
or copy the reference information returned by citation('papaja') to
your BibTeX file:
@Manual{,
title = {{papaja}: {Prepare} reproducible {APA} journal articles with {R Markdown}},
author = {Frederik Aust and Marius Barth},
year = {2022},
note = {R package version 0.1.0.9999},
url = {https://github.com/crsh/papaja},
}If you are interested in seeing how others are using papaja, you can find a collection of papers and the corresponding R Markdown files in the manual.
If you have published a paper that was written with papaja, please add the reference to the public Zotero group yourself or send us to me.
To ensure mid- to long-term computational reproducibility we highly recommend conserving the software environment used to write a manuscript (e.g. R and all R packages) either in a software container or a virtual machine. This way you can be sure that your R code does not break because of updates to R or any R package. For a brief primer on containers and virtual machines see the supplementary material by Klein et al. (2018).
Docker is the most widely used containerization approach. It is open source and free to use but requires some disk space. CodeOcean is a commercial service that builds on Docker, facilitates setting up and sharing containers and lets you run computations in the cloud. See the papaja manual on how to get started using papaja with Docker or CodeOcean and our Docker workflow tailored for easy use with papaja.
Like papaja and want to contribute? We highly appreciate any
contributions to the R package or its documentation. Take a look at the
open issues if you need
inspiration. There are many additional analyses that we would like
apa_print() to support. Any new S3/S4-methods for this function are
always appreciated (e.g., factanal, fa, lavaan). For a primer on
adding new apa_print()-methods, see the getting-started-vignette:
vignette("extending_apa_print", package = "papaja")Before working on a contribution, please review our brief contributing guidelines and code of conduct.
By now, there are a couple of R packages that provide convenience functions to facilitate the reporting of statistics in accordance with APA guidelines.
- apa: Format output of statistical tests in R according to APA guidelines
- APAstats: R functions for formatting results in APA style and other stuff
- apaTables: Create American Psychological Association (APA) Style Tables
- rempsyc: Convenience functions for psychology
- sigr: Concise formatting of significances in R
If you are looking for other journal article templates, you may be interested in the rticles package.