Master Thesis Project: Neurodegeneration identification in Parkinson's Disease with Deep learning models using 3T quantitative MRI maps
MAIA: Erasmus Mundus Joint Master Degree in Medical Imaging and Applications
This repository contains all the code and analysis notebooks for the final master's thesis project (see Report), developed at the Danish Research Centre for Magnetic Resonance in Copenhagen, Denmark. The project includes three sections:
- Dataset creation and pre-processing [/preproc]. Containing MATLAB scripts to generate quantitative MRI maps (MTsat, R1, R2*, and PD*) from the multiparametric (MPM) protocol, using the hMRI toolbox, and the bash scripts to run Synthseg parcellation and MNI PD25 atlas propagation (ANTs).
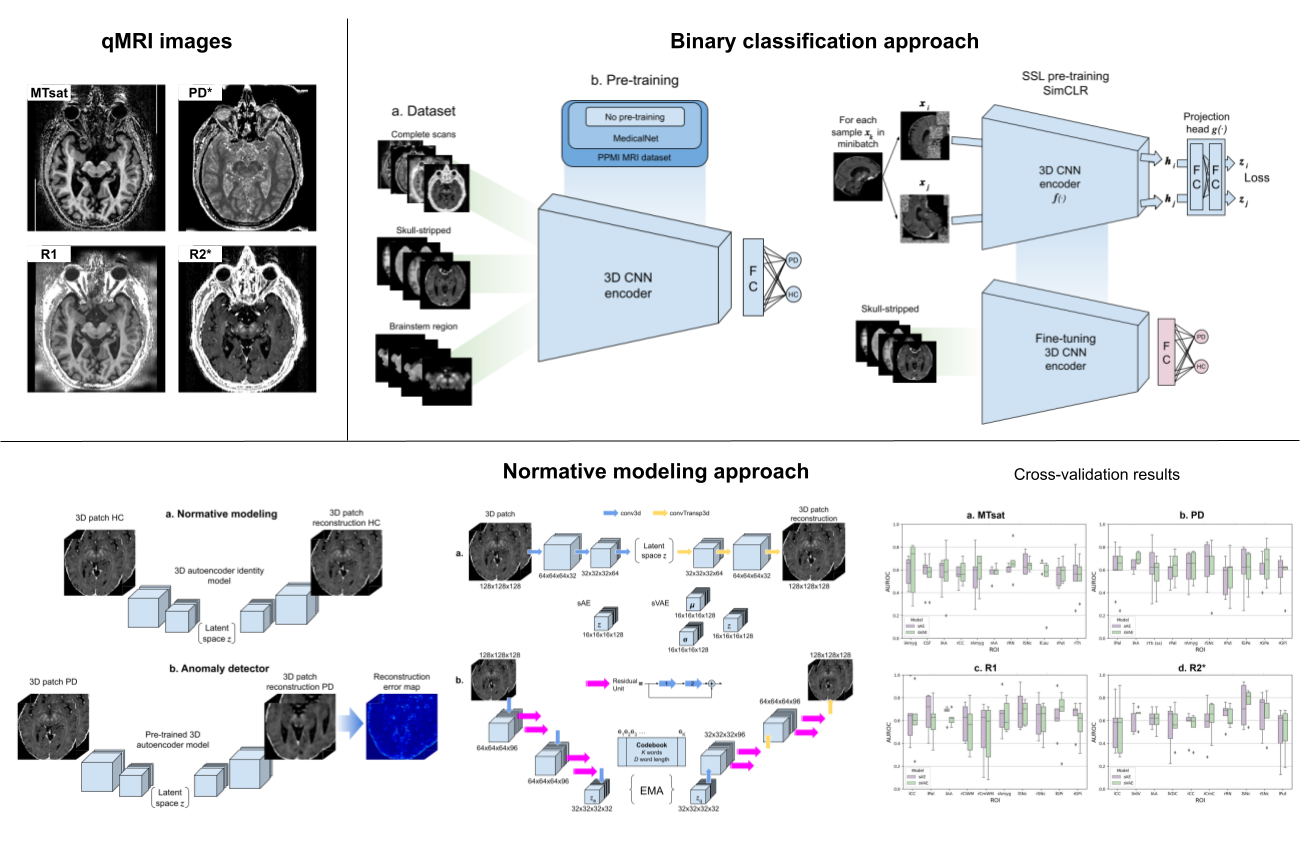
- Binary classification strategy[/bin_class]. Containing Python scripts and notebooks for models' training and inference using Pytorch Lightning. As well as the explainability scripts using XAI algorithms and final data analysis.
- Normative modeling strategy[/norm_mod]. Containing Python scripts and notebooks for models' training, inference (image reconstruction), reconstruction error maps generation, and data analysis.
Disclaimer. The purpose of these scripts and notebooks is to show the reader the implementation of the tested strategies. This project is not structured to be replicated because of data protection laws (dataset and model weights).
Start by creating a new conda environment
conda update -n base -c defaults conda &&
conda env create -f environment_short.yml &&
conda activate pdenv &&Note. The enviornment contains many libraries, so it might consume a lot of time and memory to install them. For reference of the complete list, please see environment.yml