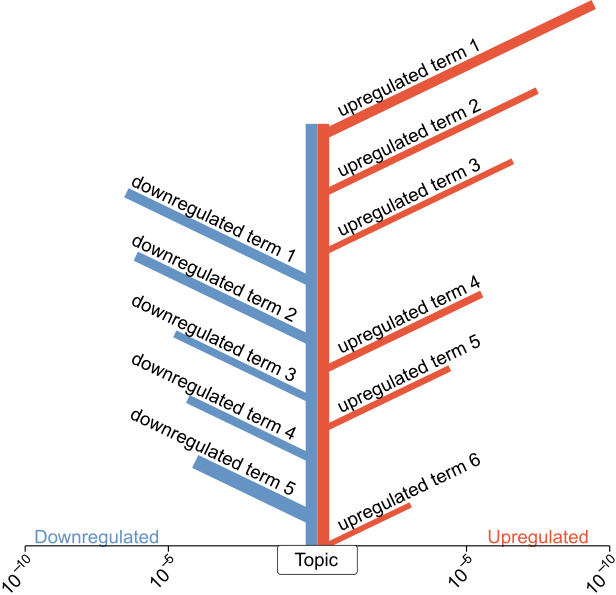
Tree Visualization for Gene Set Enrichment Results.
This package was created to provide an alternative to the omni-present horizontal bar plots used for the visualization of Gene Ontology enrichment results. Enrichment results are shown as branches of a "tree" with the length of the branches scaling with negative decimal logarithm of the enrichment p value while the branch thickness represents the number of genes in the query matching this term. The "ground" shows the axis for the p value. A defining feature of this plot is that two result sets can be presented together, quite literally "back to back", as the two sides of the tree with customizable colors. This works great for use cases such as results for different ontologies (BP, MF, CC) or to display enrichment results for down- and upregulated genes from a differential expression analysis.
Example:
Example use in publication:
Impaired Amino Acid Transport at the Blood Brain Barrier Is a Cause of Autism Spectrum Disorder. Cell, 2016.
Tărlungeanu DC, Deliu E, Dotter CP, Kara M, Janiesch PC, Scalise M, Galluccio M, Tesulov M, Morelli E, Sonmez FM, Bilguvar K, Ohgaki R, Kanai Y, Johansen A, Esharif S, Ben-Omran T, Topcu M, Schlessinger A, Indiveri C, Duncan KE, Caglayan AO, Gunel M, Gleeson JG, Novarino G.