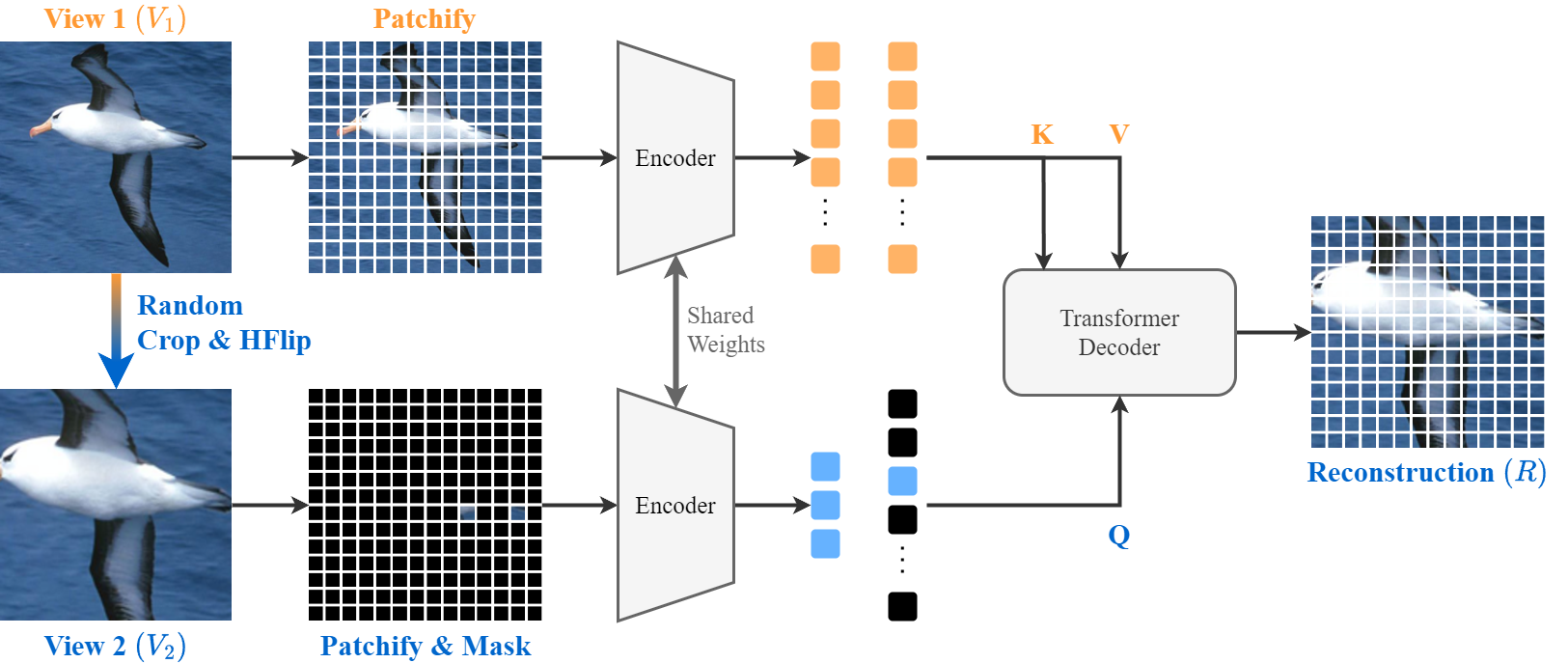
PyTorch implementation of CropMAE, introduced in Efficient Image Pre-Training with Siamese Cropped Masked Autoencoders, and presented at ECCV 2024.
| Dataset | mIoU | PCK@0.1 | Download | |
|---|---|---|---|---|
| ImageNet | 60.4 | 33.3 | 43.6 | link |
| K400 | 58.6 | 33.7 | 42.9 | link |
Run an interactive demo of CropMAE in the cloud using Colab Notebooks, or locally with the Notebook Demo.
Create a virtual environment (e.g., using conda or venv) with Python 3.11 and install the dependencies:
conda create --name CropMAE python=3.11
conda activate CropMAE
python -m pip install -r requirements.txtThis section assumes that you want to run CropMAE with default parameters. You can run python3 train_cropmae_in.py -h to have a complete list of possible parameters that you can change.
To start the training on a single GPU, you just have to provide the path to your dataset (typically ImageNet):
python train_cropmae_in.py --data_path=path/to/imagenet/folderWe provide a script to start the training on a cluster of GPUs using slurm. Modify the scripts/train_cropmae_in.sh with your slurm configuration (e.g., account name) and the parameters you want to use, and start the training with:
cd scripts && sbatch train_cropmae_in.shDownload the DAVIS, JHMDB, and VIP datasets.
The downstreams/propagation/start.py script can be used to evaluate a checkpoint on the DAVIS, JHMDB, and VIP datasets. Run the following command to have an overview of the available parameters:
python3 -m downstreams.propagation.start -hFor example, to evaluate a checkpoint on the DAVIS dataset with the default evaluation parameters (i.e, the ones used in the paper), you can use the following command:
python3 -m downstreams.propagation.start --davis --checkpoint=path/to/checkpoint.pt --output_dir=path/to/output_dir --davis_file=path/to/davis_file --davis_path=path/to/davis_pathThis will create the folder path/to/output_dir/davis and evaluate the checkpoint path/to/checkpoint.pt on DAVIS. The results, both quantitative and qualitative, will be saved in this folder, printed to the standard output stream, and reported on Weights & Biases if enabled.
If you use our code or find our results helpful, please consider citing our work:
@article{Eymael2024Efficient,
title = {Efficient Image Pre-Training with Siamese Cropped Masked Autoencoders},
author = {Eyma{\"e}l, Alexandre and Vandeghen, Renaud and Cioppa, Anthony and Giancola, Silvio and Ghanem, Bernard and Van Droogenbroeck, Marc},
journal = {arXiv:2403.17823},
year = {2024},
}- Our code is based on the official PyTorch implementation of MAE.
- The evaluation code is based on videowalk.