Winston Wang, Elisa Liu, Alex Lu
We will perform automated classification on dermoscopy images obtained from phase 3 of ISIC 2017: Skin Lesion Analysis Towards Melanoma Detection.
We will represent each image as a combination of textural features, and color information encoded in a lower dimensional space. A support vector machine will be used to classify dermoscopy images in a large dataset as melanoma or benign nevus. Leave-one-out CV will be performed over our initial training set of 2000 dermoscopy images, and final validation will be performed using a hold-out test set.
We chose SVM for its ease of use and its robustness towards overfitting. At the time, we did not consider its sensitivity to class balance and the difficulties that we would encounter with handling large numbers of features.
We chose to extract texture and color features with the understanding that both have performed exceptionally in literature -- however, we were concerned about the heterogeneous size of input images, and as a result, transitioned to L1-normalized count vectors. This turned out to be a bad decision.
The dependencies for running our support vector classifier:
numpy
scikit-learn
scikit-image
scipy
matplotlib
pandas
seaborn
The main can be run with the usage:
python mainboy.py <path to ground truth> <path to metadata> <path to image set> From scratch, the feature extraction takes about 2 hours on my machine; intermediate features from all 2000 images have been stored in folder intermediates, and can be moved to the top level if desired. If this is done, features will be read from file and the SVM will be tuned.
We also have an implementation of a not-really-well-trained convolutional neural net. For this, in addition to the above, you need keras.
The cnn can be run assigning path variables image_dir and truth_file in convnet.py before calling
python convnet.pyThese are performed within the Data Visualizations iPython notebook. See the Visualizations folder for results.
As of 12/8: 81.3% accuracy on a random subsample of images (500+) with texture and color features, using linear kernel with class_weight={1:3.5} and and C = 3.0 (?)
It took us a long time to tune it, but here we have a little bit of gridsearch:
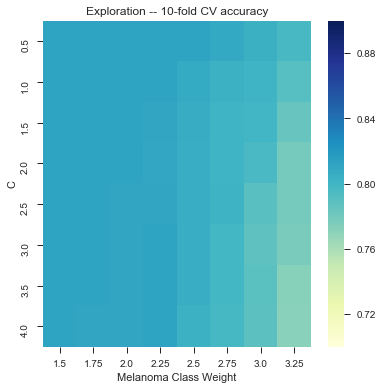
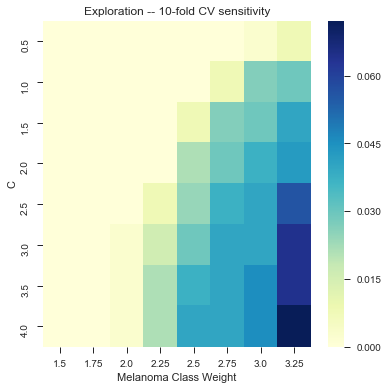
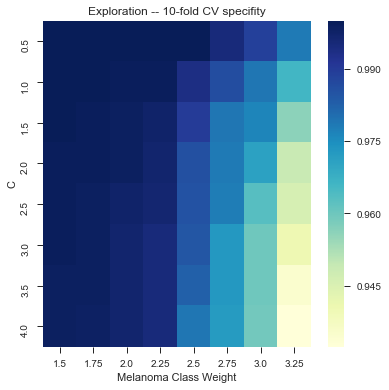
Wew lad we should have used a CNN.
The images within the dataset are of different sizes and aspect ratios, making it difficult to represent per-pixel features exactly. Rather than manual or automated cropping to achieve uniform sizes, textural features are represented as a counts to generate unit 26-dimensional vectors.
The presence of hair and other foreign bodies is a significant issue in dermoscopic analysis.

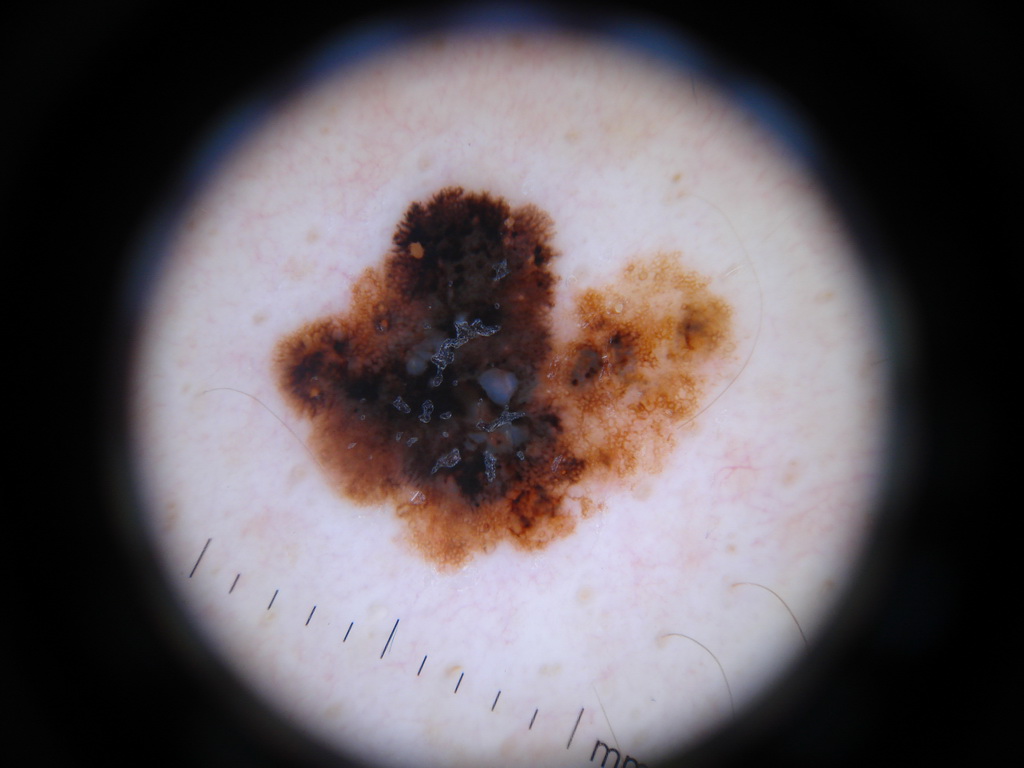
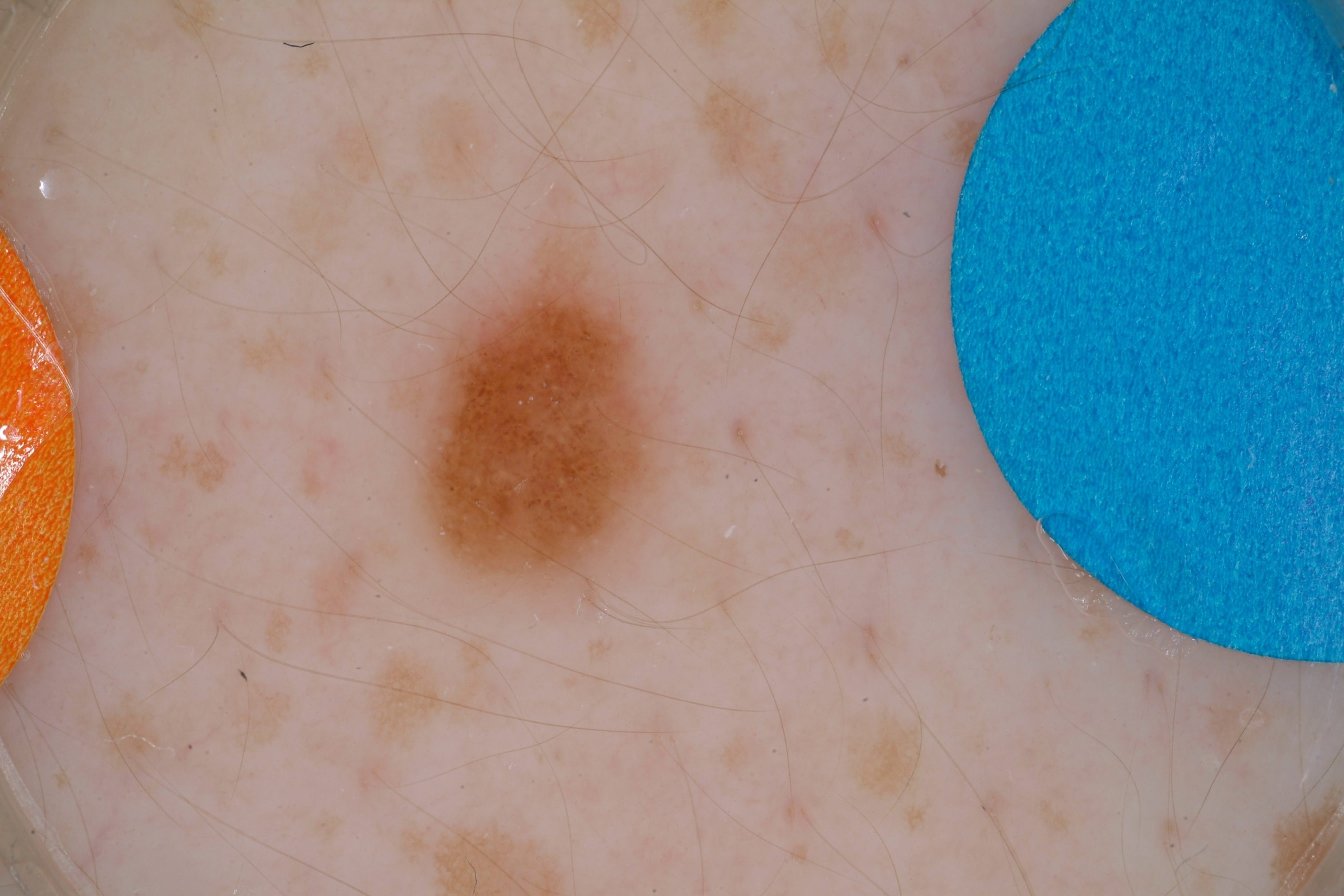
We have chosen to include these images regardless and see how well our classifier performs -- update as of 12/8/2017, 9:21PM : did not do well LOL.
We did the same feature -> count conversion for color features. The final result is an incredibly sparse series of vectors which have very little variance -- this makes it really difficult to do the machine learning, unfortunately. See:
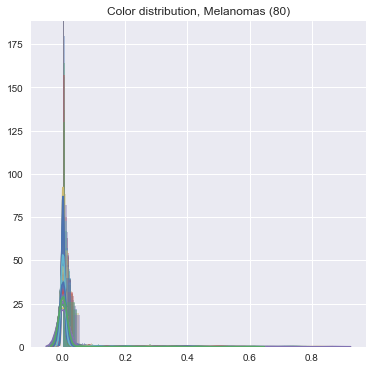
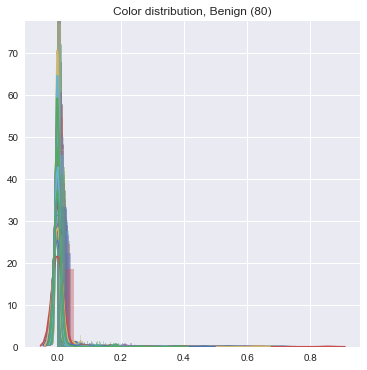
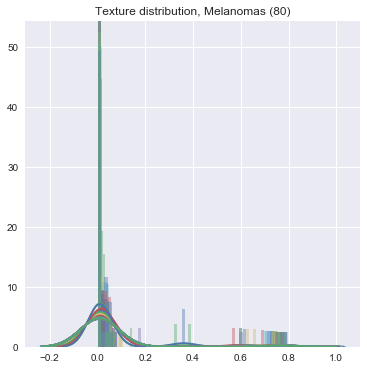
- Turn it in!