Graphical user interface that aligns two protein sequences with the selected substitution matrix using a simplified version of the needleman-wunsch algorithm. It is written in Python using the tkinter module for the GUI.
This is an improvement of my prot_align script that supports the use of user provided substitution matrices.
To launch the application in windows, open a Command prompt window in the folder of the frontend.py file and type:
py frontend.pyTo lauch the application in linux or MAC OS X, open a terminal in the folder of the frontend.py file and type:
python3 frontend.pyor (make sure you made frontend_unix.py executable with chmod)
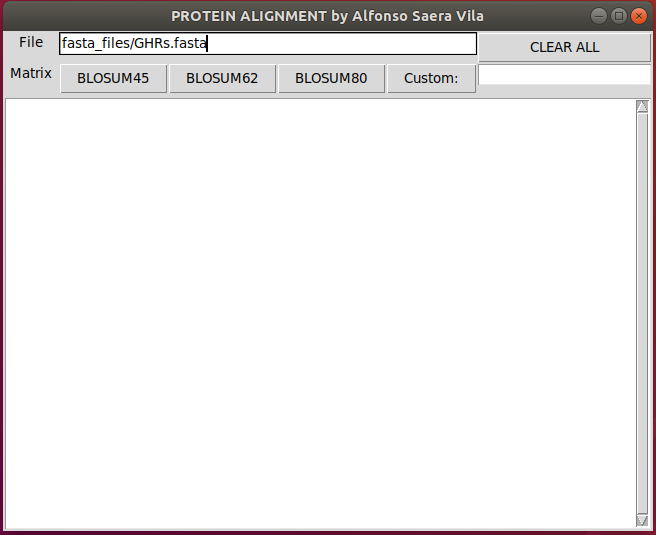
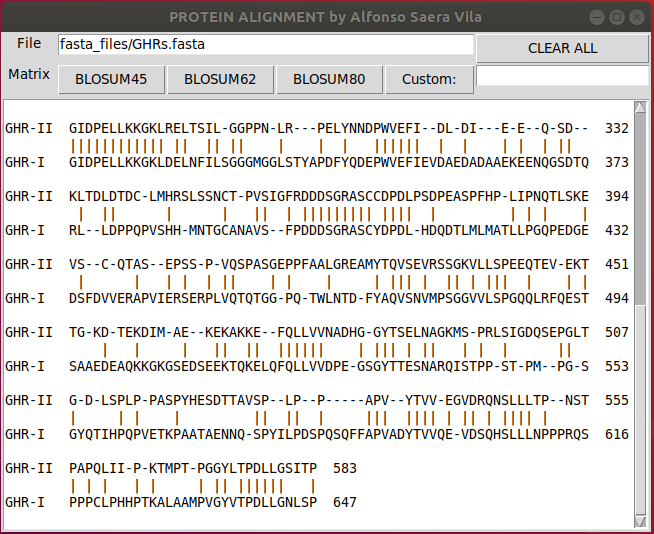
./frontend_unix.pyThen write the path of a Multi-FASTA file (the first 2 sequences will be used)
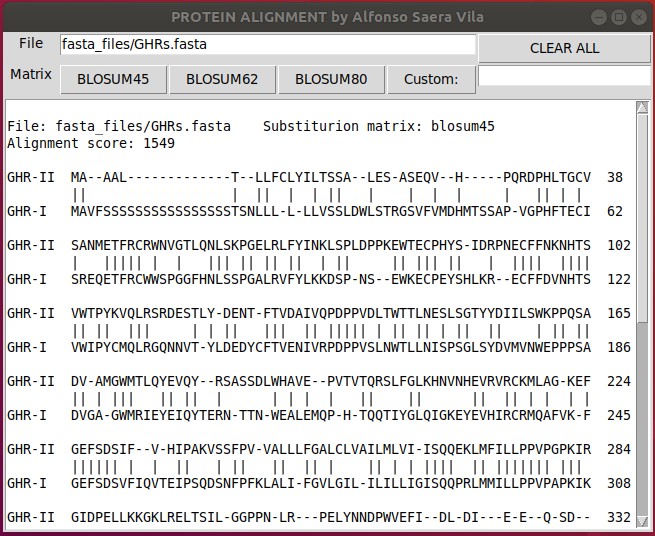
and click on the desired Blosum matrix to align the protiens
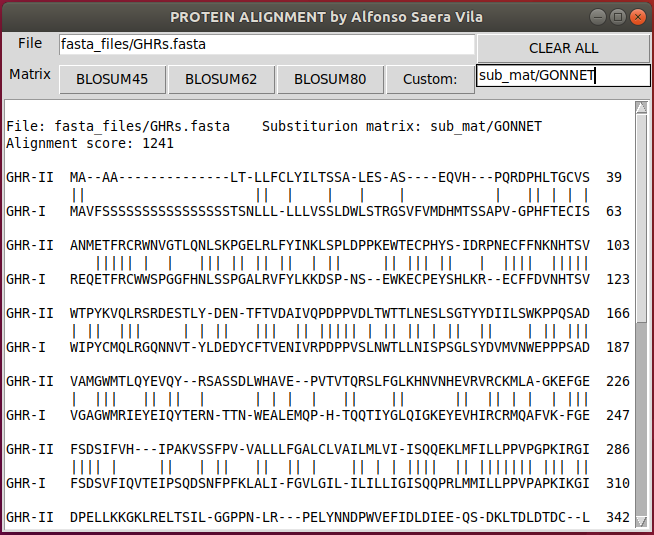
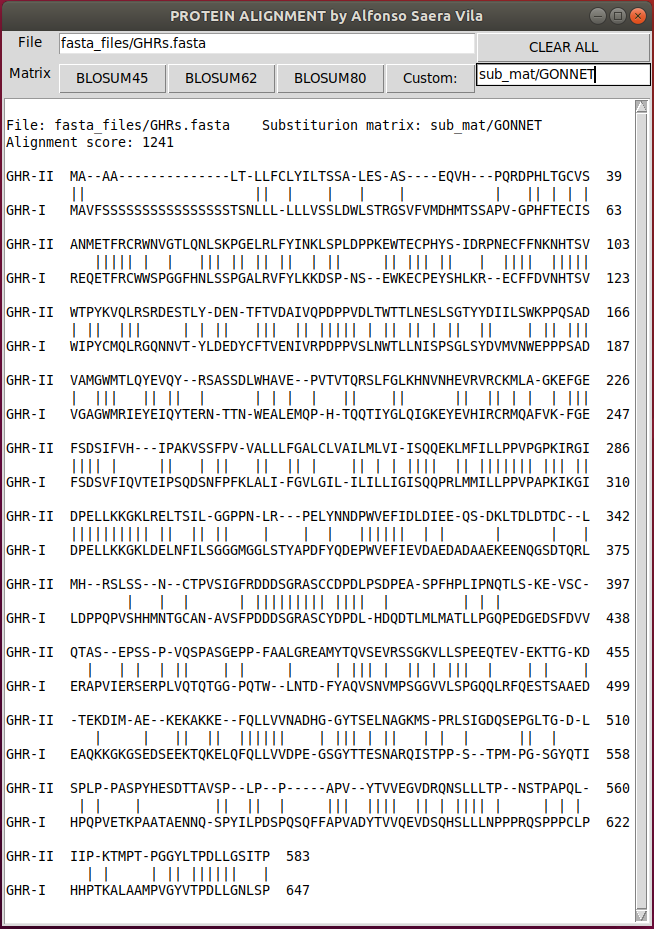
You can use any substitution matrix by writing the path to the substitution matrix file and clicking Custom: button.
Custom substitution matrices must be in genbank format (or similar enough for the matparser function). The following python code can be used to download and visualize alternative substitution matrices.
import ftplib
path = 'blast/matrices/'
filename = 'BLOSUM62'
ftp = ftplib.FTP("ftp.ncbi.nih.gov")
ftp.login('anonymous', 'password')
ftp.cwd(path)
ftp.retrbinary("RETR " + filename ,open(filename, 'wb').write)
ftp.quit()
BlossumFile = open("BLOSUM62", "r")
temp = BlossumFile.read()
BlossumFile.close()
print (temp)To visualize long alignments you can scroll down
or enlarge the window
The biopython module is required to use the predefined substitution matrices.
Download the prot_align_GUI repository from github or clone it by typing in the terminal
git clone https://github.com/alfonsosaera/prot_align_GUI.git