Anatomical Invariance Modeling and Semantic Alignment for Self-supervised Learning in 3D Medical Image Analysis
[Yankai Jiang]1,3, [Mingze Sun]1,4, [Heng Cuo]1,2, [Xiaoyu Bai]1, [Ke Yan]1,2, [Le Lu]1, [Minfeng Xu]1,2
1 DAMO Academy, Alibaba Group, 2 Hupan Lab,
3 College of Computer Science and Technology, Zhejiang University,
4 Tsinghua Shenzhen International Graduate School, Tsinghua-Berkeley Shenzhen Institute, China
ICCV, 2023, oral
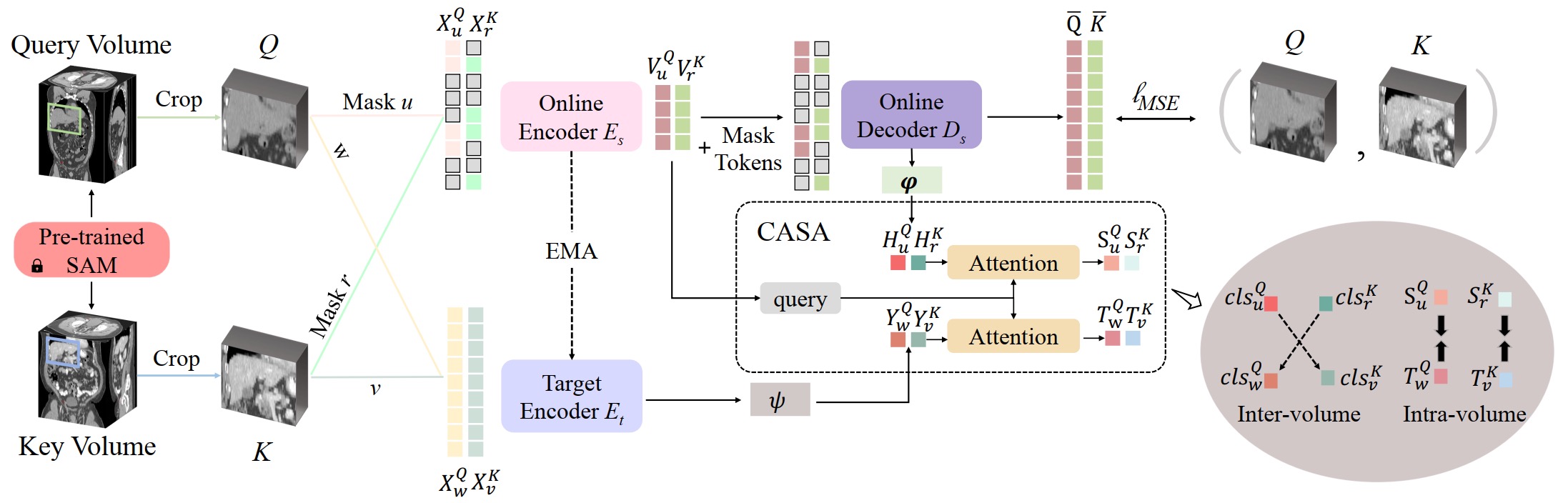
This repository contains the code for Alice (Anatomical Invariance Modeling and Semantic Alignment for Self-supervised Learning in 3D Medical Image Analysis). The architecture of Alice is illustrated below:
In Alice, a conditional anatomical semantic alignment (CASA) module is proposed to match the most related high level semantics between the crafted contrastive views. An overview of the CASA module is presented in the following:
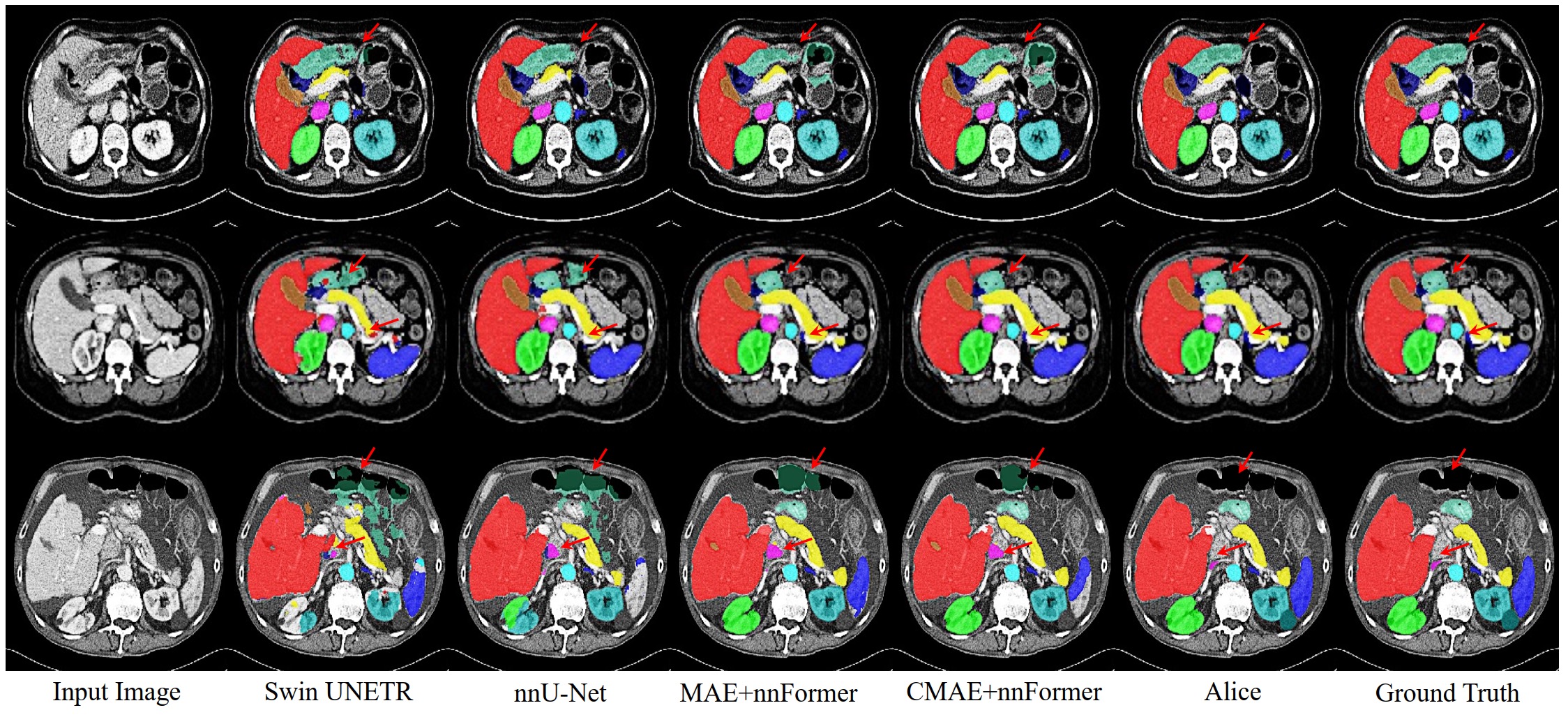
Qualitative visualizations of segmentation results:
Dependencies can be installed using:
pip install -r requirements.txt
pip install -U openmim
mim install mmcv==1.4.7
pip install mmcv-full==1.4.7 -f https://download.openmmlab.com/mmcv/dist/cu102/torch1.9/index.html
mim install mmdet==2.20The following datasets were used for pre-training (2,000 unlabeled CT scans) in our paper.
- Fast and Low-resource semi-supervised Abdominal oRgan sEgmentation in CT (FLARE 2022) (Link)
Within each dataset folder, the following structure is expected:
Dataset_FLARE2022/
├── imagesTr
└── imagesTs
You should create a json containing this dataset like pretrain_all.json. In json the data path should be '.../Dataset_FLARE2022/Case_00001_0000.nii.gz' You can use create_json.ipynb to create the json file.
Here is an example of the dataset folder, which were used for pre-training:
PreTrain/Dataset_FLARE2022/
├── dataset.json
├── imagesTr
│ ├── Case_00001_0000.nii.gz
│ ├── ...
│ ├── ...
└── imagesTs
├── ...
├── ...
├── ...
You should create a json containing this dataset like ./datasets/pretrain_all.json. You can use create_json.py to create the json file.
- Generate embeddings with a pre-trained SAM [1] model for your data. We utilize a pre-trained SAM [1] model, which performs self-supervised universal landmark detection to locate the same body part in different volumetric medical images, then we use a default input volume crop size to generate respective views of consistent anatomies.
First, clone all the codes and pre-trained weights from SamV2. Create a new virtual environment (strongly recommended) and install all the required dependencies for this repository.
Then, find misc/lymphnode_preprocess_crop_multi_process.py, change the path which saves your ".nii files", add path for generated "resampled .nii files" with masks and the path of a ".csv" file contains all saved .nii files name (index). Then run "python misc/lymphnode_preprocess_crop_multi_process.py". (You can also refer to ./sam/sam_preprocess.py in our repository.)
Next, find configs/sam/sam_NIHLN.py in [SamV2] repository and set the correct path containing your dataset.
data = dict(
test=dict(
data_dir=dlt_eval_data_root,
index_file=anno_root + 'dlt.csv',
pipeline=test_pipeline,
), )
#Also, set the output_embedding = False in the test_cfg section:
test_cfg=dict(
save_path='/data/sdd/user/results/result-dlt/',
output_embedding=False) # False: save the embeeding to the save_path; True: return the embdding
Finally, run dist_tesh.sh #PATH_TO_CONFIG #NUM_GPU. The embeddings for all cases will be saved in the save_path.
You can also refer to ./sam/sam_AbdomenAtlas.py and ./sam/sam_dist_test.sh in our repository, which show our modifications to the configs/sam/sam_NIHLN.py file and the dist_tesh.sh file from [SamV2] repository. You can use these script examples to process your own dataset.
- Distributed Multi-GPU Pre-Training.
Change main settings in main-DDP.py:
--data_dir = ".../../PreTrain/Dataset_FLARE2022/" ## The directory where the raw data are stored.
--json_list = "pretrainset_all.json" ## The directory where the json_file generated by create_json.py are stored.
--output_dir = "./results/" ## Path to save logs and checkpoints.
--embed_dir = "SAM Embedding" ## The directory where the embeddings generated by [SamV2] repository are stored.
--checkpoint_file = "SAM pretrain checkpoint" ## The directory where the checkpoints of a pre-trained SAM [1] model are stored. you can find the checkpoint in [SamV2] repository. Here we also provide it in our repository: ./checkpoints_SAM/SAM.pth
The following command is used to pre-train Alice on 8 X 80G A100 GPUs:
python3 -m torch.distributed.launch --nproc_per_node=8 main-DDP.py
--batch_size=8 --lr=5e-5- Finetuning.
After pretraining with Alice, you can utilize the script extract_backbone_weights.py to extract the weights from either the 'student' or 'teacher' branch for your downstream tasks.
example: python extract_backbone_weights.py /mnt/workspace/jiangyankai/Alice_code/results/final-ddp/checkpoint0030.pth /mnt/workspace/jiangyankai/pretrained_weights/checkpoint.pth
Note that the objectives of self-supervised training may differ significantly from those of your downstream tasks. Therefore, you may need to fine-tune your model on your specific downstream dataset.
We have recently applied Alice's pre-training techniques to the dataset from the paper 'AbdomenAtlas-8K: Annotating 8,000 CT Volumes for Multi-Organ Segmentation in Three Weeks' (Qu et al. published in NeurIPS 2023). You will find our pre-trained parameters in the './results' directory.
If you find this repository useful, please consider citing Alice paper:
@inproceedings{jiang2023anatomical,
title={Anatomical Invariance Modeling and Semantic Alignment for Self-supervised Learning in 3D Medical Image Analysis},
author={Jiang, Yankai and Sun, Mingze and Guo, Heng and Bai, Xiaoyu and Yan, Ke and Lu, Le and Xu, Minfeng},
booktitle={Proceedings of the IEEE/CVF International Conference on Computer Vision},
pages={15859--15869},
year={2023}
}
This code is based on the implementations of MAE, SimMIM, SamV2, SwinTransformer, iBOT, and DINO. We deeply appreciate all these exceptional and inspiring works!
[1]: Yan K, Cai J, Jin D, et al. SAM: Self-supervised learning of pixel-wise anatomical embeddings in radiological images[J]. IEEE Transactions on Medical Imaging, 2022, 41(10): 2658-2669.