The official implementation of Alina Muliak's Thesis under the supervision of Amar Basu submitted in fulfillment of the requirements for the degree of Bachelor of Science in the Department of Computer Sciences and Information Technologies Faculty of Applied Sciences.
This repository is the official implementation of Bachelor Thesis on Deriving Biometrics for Orthostatic Hypotension (OH).
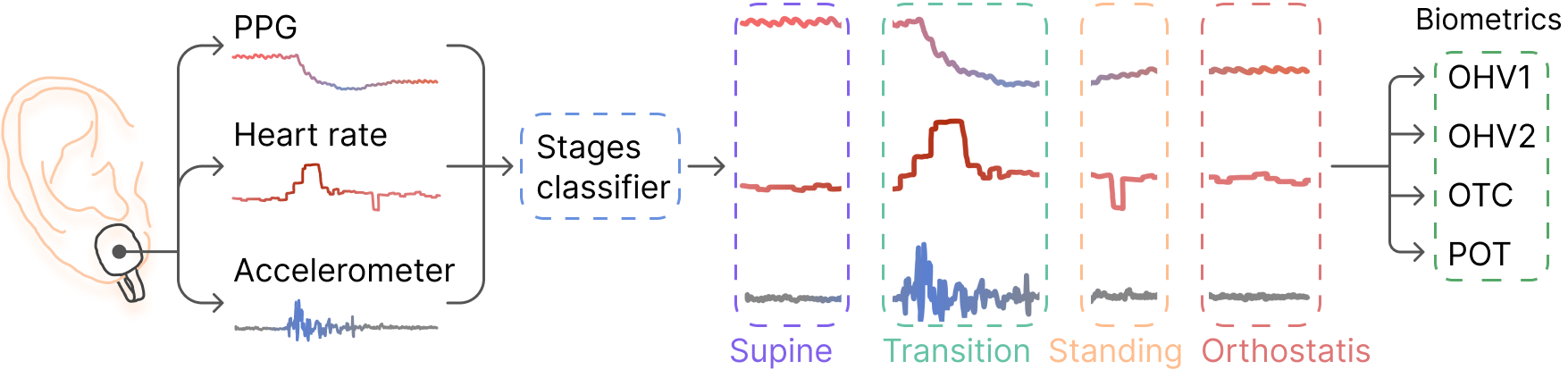
The proposed approach involves classifying the signals during stand-up tests into four phases: supine, transition,
standing, and orthostasis. The classified phases are then used to calculate the biometrics, which indicate the severity
of the OH in a patient.
To run and test our code, follow the steps below.
add info about python version
git clone https://github.com/alinamuliak/deriving-biometrics-for-oh.gitcd deriving-biometrics-for-ohpip install virtualenv, ifvirtualenvis not installed yetvirtualenv venvsource venv/bin/activateon Unix system;venv\Scripts\activateon Windowspip install -r requirements.txt- Navigate to git-lfs.com and click Download.
git lfs installgit lfs pull
If you have CUDA available on your laptop, download torch from the PyTorch official site,
choosing your specific settings. If the CUDA is available, it will be used by default. Otherwise, the CPU will be used.
For instance, using Windows with CUDA 11.8, execute:
pip3 install torch torchvision torchaudio --index-url https://download.pytorch.org/whl/cu118
For each type of the models, a separate training script is used, because of lot different parameters between the models.
| Model | Script name |
|---|---|
| CNN | train_cnn.py |
| LSTM | train_lstm.py |
| Hybrid | train_hybrid.py |
To see more detailed description about each parameter, run python train_<model>.py --help or python train_<model>.py -h.
All the parameters are optional and if not provided, the default values from the thesis will be used.
To train the CNN model, run this command:
python train_cnn.py [-h] [--batch_size BATCH_SIZE] [--n_conv_blocks N_CONV_BLOCKS]
[--out_channels OUT_CHANNELS] [--dropout DROPOUT] [--learning_rate LEARNING_RATE]
[--weight_decay WEIGHT_DECAY] [--num_epochs NUM_EPOCHS] [--verbose] [--model_name MODEL_NAME]
To train the LSTM model, run this command:
python train_lstm.py [-h] [--batch_size BATCH_SIZE] [--bidirectional] [--num_layers NUM_LAYERS]
[--hidden_size HIDDEN_SIZE] [--dropout DROPOUT] [--learning_rate LEARNING_RATE]
[--weight_decay WEIGHT_DECAY] [--num_epochs NUM_EPOCHS]
[--verbose] [--model_name MODEL_NAME]
To train the hybrid model, run this command:
python train_hybrid.py [-h] [--n_conv_blocks N_CONV_BLOCKS] [--cnn_out_channels CNN_OUT_CHANNELS]
[--cnn_dropout CNN_DROPOUT] [--lstm_bidirectional] [--lstm_num_layers LSTM_NUM_LAYERS]
[--lstm_hidden_size LSTM_HIDDEN_SIZE] [--lstm_dropout LSTM_DROPOUT]
[--learning_rate LEARNING_RATE] [--weight_decay WEIGHT_DECAY] [--num_epochs NUM_EPOCHS]
[--verbose] [--model_name MODEL_NAME]
To evaluate the model on test dataset, run:
python eval.py [-h] --model_type {cnn,lstm,hybrid} --chkpt_path CHECKPOINT_PATH --batch_size BATCH_SIZE [--device {cuda,cpu,mps}] [--save_plots_to SAVE_PLOTS_TO]
To evaluate models, implemented in the Thesis, specify the checkpoint path of the predefined models
located in models directory along with the batch size specified below. For instance,
python eval.py --model_type hybrid --chkpt_path models/hybrid.pt --batch_size 32
After evaluation is done, a table containing accuracy, f1-score, MAE and MPE will be printed.
You can find pretrained models in models directory, where the name of the file corresponds with the model used.
- CNN trained using the following parameters: 2 convolution blocks, kernel size of 32, dropout of 0.5, learning rate of 5e-05, weight decay of 0.0005 and 298 epochs.
- UniLSTM trained using the following parameters: 2 layers with hidden size 231, dropout of 0.583, learning rate of 0.0005, weight decay 9e-06 and 1110 epochs.
- BiLSTM trained using the following parameters: 2 layers with hidden size of 200, dropout of 0.65, learning rate of 0.001, weight decay of 7.5e-05 and 899 epochs.
- Hybrid trained using the following parameters: 2 convolution blocks, CNN kernel size of 32, CNN dropout of 0.388, Bidirectional LSTM, 3 LSTM layers with hidden size of 174, LSTM dropout of 0.269, learning rate of 0.00973, weight decay of 7e-05 and 250 epochs.
The achieved performance metrics for each model are presented in the tables below. The best model was hybrid, which achieved F1-score of 91.9% on the test dataset.
| Model | Accuracy | F1-score |
|---|---|---|
| CNN | 71.4% | 71.1% |
| UniLSTM | 86.0% | 86.9% |
| BiLSTM | 84.6% | 85.8% |
| Hybrid model | 91.3% | 91.8% |
| Model | OHV1 MAE [a. u.] | OHV2 MAE [a. u.] | OTC MAE [sec] | POT MAE [bpm] |
|---|---|---|---|---|
| CNN | 315.44 | 441.79 | 8.43 | 15.84 |
| UniLSTM | 303.75 | 179.28 | 6.82 | 21.14 |
| BiLSTM | 220.07 | 63.0 | 21.24 | 5.94 |
| Hybrid | 54.32 | 64.5 | 3.39 | 11.88 |
| Model | OHV1 MPE | OHV2 MPE | OTC MPE | POT MPE |
|---|---|---|---|---|
| CNN | 130.7% | 802.77% | 40.98% | 64.74% |
| UniLSTM | 350.71% | 88.48% | 38.37% | 112.28% |
| BiLSTM | 439.48% | 4.4% | 162.0% | 25.56% |
| Hybrid | 9.76% | 16.94% | 14.8% | 53.48% |
- Alina Muliak
- Amar Basu
💡 README template from here.