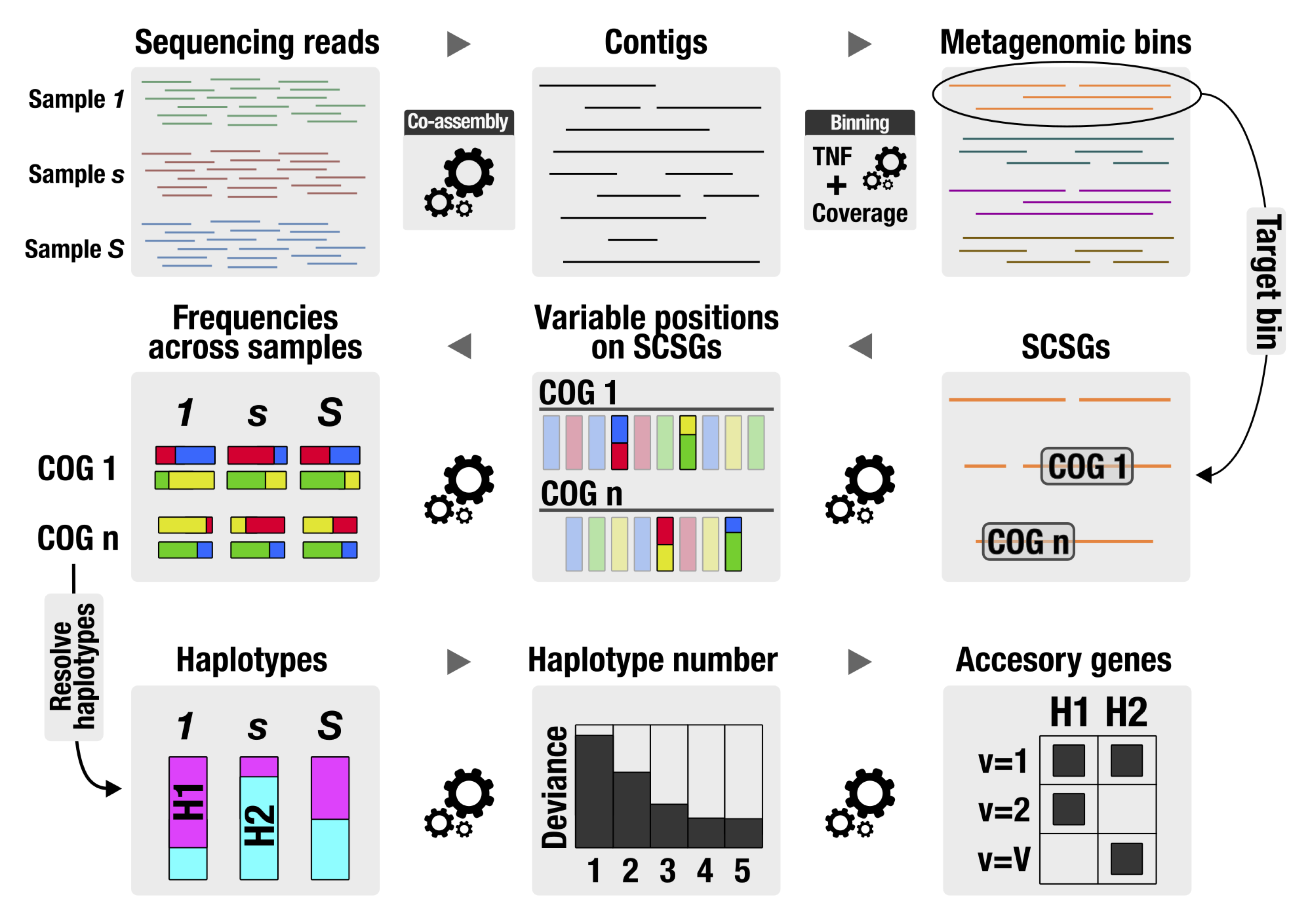
The DESMAN pipeline is described in the bioRxiv preprint.
##Installation
To install simply type:
sudo python ./setup.py install
These items are prerequisities for the installation of desman:
- python v2.7.
- *gcc
- *gsl
The installation procedure varies on different systems, and described in this README is only how to proceed with a linux (ubuntu) distribution.
The first item, python v2.7.*, should be installed on a modern Ubuntu distribution. A c-compiler, e.g. gcc, is needed to compile the c parts of concoct that uses the GNU Scientific Library gsl. For linux (ubuntu) this is installed through:
sudo apt-get install build-essential libgsl0-dev
For convenience we also recommend adding the scripts directory to your path:
export PATH=$HOME/myinstalldir/DESMAN/scripts:$PATH
Obviously replacing myinstalldir as appropriate and adding this to your .bash_profile file.
##Simple exampleTo illustrate the actual strain inference algorithm we will start with a simple example using base frequencies that have been pre-prepared. Below we also give a complete example including pre-processing. The starting point for a Desman analysis is a csv file with base frequencies e.g.:
Strain mock community frequencies for COG0015
This has the following format:
Contig,Position,SampleName1-A,SampleName1-C,SampleName1-G,SampleName1-T,...,SampleNameN-A,SampleNameN-C,SampleNameN-G,SampleNameN-T
where SampleName1,...,SampleNameN gives the names of the different samples in the analysis. Followed by one line for each position with format:
gene name, position, freq. of A in sample 1, freq. of C in 1,freq. of G in 1,freq. of T in 1,..., freq. of A in sample N, freq. of C in N,freq. of G in N,freq. of T in N
###Finding variant positions for the test data set
The first step is to identify variant positions. This is performed by the desman script Variant_Filter.py. Start assuming you are in the DESMAN repo directory by making a test folder.
mkdir test
cd test
Then run the example data file which corresponds to a single COG from the mock community data set described in the manuscript. This COG0015 has 933 variant positions. The input file is in the data folder. We run the variant filtering as follows:
python ../desman/Variant_Filter.py ../data/contig_6or16_genesL_scgCOG0015.freq -o COG0015_out -p
The variant filtering has a number of optional parameters to see them run:
python ../desman/Variant_Filter.py -h
They should all be fairly self explanatory. We recommend always using the the '-p' flag for one dimenisonal optimisition of individual base frequencies if it is not too time consuming. The '-o' option is a file stub all output files will be generated with this prefix. A log file will be generated 'COG0015_out_log.txt' and output files:
-
COG0015_outp_df.csv: This gives p-values for each position.
-
COG0015_outq_df.csv: This gives q-values for each position.
-
COG0015_outr_df.csv: This gives log-ratio statistics for each position.
-
COG0015_outsel_var.csv: This is the file of selected variants.
-
COG0015_outtran_df.csv: A matrix of estimated error rates.
###Inferring haplotypes and abundances for the test data set
Having found the variant positions we will now the run the program for inferring haplotypes and their abundance:
desman COG0015_outsel_var.csv -g 5 -e COG0015_outtran_df.csv -o COG0015_out_g5 -i 50
These parameters specify the variants file. Then number of haplotypes as five '-g 5', an initial estimate for the error transition matrix taken from the variant detection '-e COG0015_outtran_df.csv', an output directory '-o COG0015_out_g5' and the number of iterations, '-i 50'. The program takes the selected variants and infers haplotypes and their abundances using the Gibbs sampler given the assumption that five strains are present. All output files will be generated in the directory COG0015_out_g5. Once the program has finished running a few minutes on a typical computer it will generate the following files inside the output directory:
-
log_file.txt: This logs the progress of the program through the three stages: NTF initialisation, 'burn-in' Gibbs sampler and the sampling itself.
-
Eta_star.csv: Prediction for error transition matrix (rows true bases, columns observed probabilities) taken from sample with largest log posterior.
-
Eta_mean.csv: Prediction for error transition matrix (rows true bases, columns observed probabilities) as the posterior mean calculated over all samples.
-
Filtered_Tau_star.csv: Prediction for strain haplotypes. Each row of comma separated file contains:
gene name, position, haplotype1-A, haplotype1-C, haplotype1-G, haplotype1-T,..., haplotypeG-A, haplotypeG-C, haplotypeG-G, haplotypeG-T
where 1 indicates the base present in that haplotype at that position.
-
Tau_mean.csv: As above but the posterior mean. This can lead to non-discrete haplotypes. This can be viewed as posterior probability of assignments. Prior to analysis results should be discretised.
-
Gamma_star.csv: This gives the relative frequency of each haplotye in each sample using the sample with largest log posterior. One row for each sample.
-
Gamma_mean.csv: As above but posterior mean.
-
Selected_variants.csv: Variants used for strain calling if filtering applied.
-
fit.txt: Statistics evaluating fit as number of haplotypes, number of non-degenerate haplotypes inferred, log maximum posterior probability, mean posterior deviance (-2.0*Log likelihood)
-
fitF.txt: Statistics evaluating fit of assigned haplotypes (not in random subsample) as number of haplotypes, number of non-degenerate haplotypes inferred, log maximum posterior probability, mean posterior deviance (-2.0*Log likelihood). Only generated if -r option used.
#Complete example of de novo strain level analysis from metagenome data#
Read assembly, mapping and binning
To provide an in depth illustration of how to use Deman we will give a complete worked example from a subset of the synthetic community used in Quince et al. 2016. We have provided 16 samples, subsampled to 1 million reads from the 64 samples with 11.75 million reads used originally. This example is therefore more tractable but the following analysis assumes you have access to a multi-core server. We also assume that you have some standard and not so standard sequence analysis software installed:
-
megahit: A highly efficient metagenomics assembler currently our default for most studies
-
bwa: Necessary for mapping reads onto contigs
-
bam-readcount: Used to get per sample base frequencies at each position
-
[samtools] (http://www.htslib.org/download/): Utilities for processing mapped files
-
CONCOCT: Our own contig binning algorithm
-
[prodigal] (https://github.com/hyattpd/prodigal/releases/): Used for calling genes on contigs
-
[gnu parallel] (http://www.gnu.org/software/parallel/): Used for parallelising rps-blast
-
[standalone blast] (http://www.ncbi.nlm.nih.gov/books/NBK52640/): Need rps-blast
-
COG RPS database: ftp://ftp.ncbi.nih.gov/pub/mmdb/cdd/little_endian/ Cog databases
-
[GFF python parser] (https://github.com/chapmanb/bcbb/tree/master/gff)
Click the link associated with each application for installation details. To begin obtain the reads from Dropbox:
wget https://www.dropbox.com/s/l6g3culvibym8g7/Example.tar.gzRename, untar and unzip the example directory and move into it:
tar -xvzf Example.tar.gz
cd Example##Read assembly, mapping and binning##
Then assemble the reads. We recommend megahit for this:
nohup megahit -1 $(<R1.csv) -2 $(<R2.csv) -t 36 -o Assembly --presets meta > megahit.out&This will take a while so we have set megahit running on 36 threads (adjust to your system) and run in background with nohup.
We will now perform CONCOCT binning of these contigs. As explained in Alneberg et al. there are good reasons to cut up contigs prior to binning. We will use a script from CONCOCT to do this. For convenience we will create environmental variables points to the CONCOCT and DESMAN install directories:
export CONCOCT=$HOME/mypathtoConcoct/CONCOCT
export DESMAN=$HOME/mypathtoDesman/DESMAN
export DESMAN_EXAMPLE=$HOME/mypathtoDesmanExample/ExampleThen cut up contigs and place in new dir:
mkdir contigs
python $CONCOCT/scripts/cut_up_fasta.py -c 10000 -o 0 -m Assembly/final.contigs.fa > contigs/final_contigs_c10K.faHaving cut-up the contigs the next step is to map all the reads from each sample back onto them. First index the contigs with bwa:
cd contigs
bwa index final_contigs_c10K.fa
cd ..Then perform the actual mapping you may want to put this in a shell script:
mkdir Map
for file in *R1.fastq
do
stub=${file%_R1.fastq}
echo $stub
file2=${stub}_R2.fastq
bwa mem -t 32 contigs/final_contigs_c10K.fa $file $file2 > Map/${stub}.sam
doneHere we are using 32 threads for bwa mem '-t 32' you can adjust this to whatever is suitable for your machine. Then we need to calculate our contig lengths using one of the Desman scripts.
python $DESMAN/scripts/Lengths.py -i contigs/final_contigs_c10K.fa > contigs/final_contigs_c10K.lenThen we calculate coverages for each contig in each sample:
for file in Map/*.sam
do
stub=${file%.sam}
stub2=${stub#Map\/}
echo $stub
(samtools view -h -b -S $file > ${stub}.bam; samtools view -b -F 4 ${stub}.bam > ${stub}.mapped.bam; samtools sort -m 1000000000 ${stub}.mapped.bam -o ${stub}.mapped.sorted.bam; bedtools genomecov -ibam ${stub}.mapped.sorted.bam -g contigs/final_contigs_c10K.len > ${stub}_cov.txt)&
doneand use awk to aggregate the output of bedtools:
for i in Map/*_cov.txt
do
echo $i
stub=${i%_cov.txt}
stub=${stub#Map\/}
echo $stub
awk -F"\t" '{l[$1]=l[$1]+($2 *$3);r[$1]=$4} END {for (i in l){print i","(l[i]/r[i])}}' $i > Map/${stub}_cov.csv
doneand finally run the following perl script to collate the coverages across samples, where we have simply adjusted the format from csv to tsv to be compatible with CONCOCT:
$DESMAN/scripts/Collate.pl Map | tr "," "\t" > Coverage.tsvand run CONCOCT:
mkdir Concoct
cd Concoct
mv ../Coverage.tsv .
concoct --coverage_file Coverage.tsv --composition_file ../contigs/final_contigs_c10K.fa
cd ..In this case we know which contig derives from which of the 20 genomes and so we can compare the assignment of contigs to clusters with those genome assignments. To get the genome assignments we first need the strain genomes:
wget https://www.dropbox.com/s/9ozp0vvk9kg2jf0/Mock1_20genomes.fasta
mkdir AssignGenome
mv Mock1_20genomes.fasta AssignGenome/Mock1_20genomes.fasta
We need to index our bam files:
for file in Map/*mapped.sorted.bam
do
stub=${file%.bam}
stub2=${stub#Map\/}
echo $stub
samtools index $file
done
Then we run a script that extracts the mock genome ids out of the fastq ids of the simulated reads:
cd AssignGenome
python $DESMAN/scripts/contig_read_count_per_genome.py final_contigs_c10K.fa Mock1_20genomes.fasta ../Map/*mapped.sorted.bam > final_contigs_c10K_genome_count.tsv
cd ..
This file contains counts of unambiguous and ambiguous reads mapping to each of the genomes for each of the contigs. We simplify the genome names and filter these counts:
$DESMAN/scripts/MapGHeader.pl $DESMAN/complete_example/Map.txt < AssignGenome/final_contigs_c10K_genome_count.tsv > AssignGenome/final_contigs_c10K_genome_countR.tsv
Then we get assignments of each contig to each genome:
$DESMAN/scripts/LabelSMap.pl Concoct/clustering_gt1000.csv AssignGenome/final_contigs_c10K_genome_countR.tsv > AssignGenome/clustering_gt1000_smap.csv
This enables to compare the CONCOCT clusterings with these assignments:
$CONCOCT/scripts/Validate.pl --cfile=Concoct/clustering_gt1000.csv --sfile=AssignGenome/clustering_gt1000_smap.csv --ffile=contigs/final_contigs_c10K.fa
This should generate output similar too:
N M TL S K Rec. Prec. NMI Rand AdjRand
9159 9159 5.8184e+07 20 25 0.986561 0.992898 0.988337 0.998623 0.988009
The exact results may vary but the overall accuracy should be similar. We can also plot the resulting confusion matrix:
$CONCOCT/scripts/ConfPlot.R -c Conf.csv -o Conf.pdf
From this it is apparent that five clusters: D1, D9, D11, D15, and D18 represent the E. coli pangenome. In general, it will not be known a priori from which taxa a cluster derives and so not possible to link them in this way. However, in many analyses the pangenome will be contained in a single cluster or a contig taxonomic classifier could be used to determine clusters deriving from the same species. We illustrate how to do this below. In your particular run these assignments may vary and the code below must be changed accordingly.
##Taxonomic classification of contigs
There are many ways to taxonomically classify assembled sequence. We suggest a gene based approach. The first step is to call genes on all contigs that are greater than 1,000 bp. Shorter sequences are unlikely to contain complete coding sequences. The following requires that you have a Diamond formatted version of the NCBI NR on your system. To ensure compatibility with the files below this can be downloaded by:
wget http://nrdatabase.s3.climb.ac.uk/nr.dmnd
Set the environment variable NR_DMD to point to the location of this file:
export NR_DMD=$HOME/native/Databases/nr/FASTA/nr.dmnd
Then we begin by calling genes on all contigs greater than 1000bp in length.
mkdir Annotate_gt1000
cd Annotate_gt1000
python $DESMAN/scripts/LengthFilter.py -m 1000 ../contigs/final_contigs_c10K.fa > final_contigs_gt1000_c10K.fa
prodigal -i final_contigs_gt1000_c10K.fa -a final_contigs_gt1000_c10K.faa -d final_contigs_gt1000_c10K.fna -f gff -p meta -o final_contigs_gt1000_c10K.gff
cd ..
mkdir AssignTaxa
cd AssignTaxa
cp ../Annotate_gt1000/final_contigs_gt1000_c10K.faa .
diamond blastp -p 32 -d $NR_DMD -q final_contigs_gt1000_c10K.faa -a final_contigs_gt1000_c10K > d.out
diamond view -a final_contigs_gt1000_c10K.daa -o final_contigs_gt1000_c10K_nr.m8
To classify the contigs we need two files a gid to taxid mapping file and a mapping of taxaid to full lineage:
-
gi_taxid_prot.dmp
-
all_taxa_lineage_notnone.tsv
These can also be downloaded from the Dropbox:
wget https://www.dropbox.com/s/x4s50f813ok4tqt/gi_taxid_prot.dmp.gz
wget https://www.dropbox.com/s/honc1j5g7wli3zv/all_taxa_lineage_notnone.tsv.gz
The path to these files are default in the ClassifyContigNR.py script as the variables:
DEF_DMP_FILE = "/home/chris/native/Databases/nr/FASTA/gi_taxid_prot.dmp"
DEF_LINE_FILE = "/home/chris/native/Databases/nr/FASTA/all_taxa_lineage_notnone.tsv"
We calculate the gene length in amino acids before running this. Then we can assign the contigs and genes called on them:
python $DESMAN/scripts/Lengths.py -i final_contigs_gt1000_c10K.faa > final_contigs_gt1000_c10K.len
python $DESMAN/scripts/ClassifyContigNR.py final_contigs_gt1000_c10K_nr.m8 final_contigs_gt1000_c10K.len -o final_contigs_gt1000_c10K_nr -l /mypath/all_taxa_lineage_notnone.tsv -g /mypath/gi_taxid_prot.dmp
Then we extract species out:
$DESMAN/scripts/Filter.pl 8 < final_contigs_gt1000_c10K_nr_contigs.csv | grep -v "_6" | grep -v "None" > final_contigs_gt1000_c10K_nr_species.csv
These can then be used for the cluster confusion plot:
$CONCOCT/scripts/Validate.pl --cfile=../Concoct/clustering_gt1000.csv --sfile=final_contigs_gt1000_c10K_nr_species.csv --ffile=../contigs/final_contigs_c10K.fa --ofile=Taxa_Conf.csv
Now the results will be somewhat different...
N M TL S K Rec. Prec. NMI Rand AdjRand
9159 6926 4.8270e+07 56 25 0.940151 0.994404 0.969259 0.994666 0.955695
With a decrease in recall because the taxonomically classification is overestimating the diversity of organisms present. We then plot the out Conf.csv which contains species proportions in each cluster:
$CONCOCT/scripts/ConfPlot.R -c Taxa_Conf.csv -o Taxa_Conf.pdf
This confirms from a de novo approach that D1, D9, D11, D15 and D18 represent the E. coli pangenome.
##Identifying E. coli core genes
We now determine core genes single copy genes within these four clusters through annotation to COGs. First lets split the contigs by their cluster and concatenate togethers those from D1, D20, D22, and D23 into one file ClusterEC.fa. If your clustering gave different bins associated with E. coli then change the files selected below as appropriate:
Go back to the top level example directory and then:
mkdir Split
cd Split
$DESMAN/scripts/SplitClusters.pl ../contigs/final_contigs_c10K.fa ../Concoct/clustering_gt1000.csv
cat Cluster1/Cluster1.fa Cluster9/Cluster9.fa Cluster11/Cluster11.fa Cluster15/Cluster15.fa Cluster18/Cluster18.fa > ClusterEC.fa
cd ..Now call genes on the E. coli contigs.
mkdir AnnotateEC
cd AnnotateEC
cp ../Split/ClusterEC.fa .
prodigal -i ClusterEC.fa -a ClusterEC.faa -d ClusterEC.fna -f gff -p meta -o ClusterEC.gffNext we assign COGs using the CONCOCT script RPSBLAST.sh. First set location of your COG rpsblast database. Then run the CONCOCT script. This requires rpsblast and gnu parallel.
export COGSDB_DIR=~/gpfs/Databases/rpsblast_db
$CONCOCT/scripts/RPSBLAST.sh -f ClusterEC.faa -p -c 8 -r 1and extract out the annotated Cogs associated with called genes:
$DESMAN/scripts/ExtractCogs.py -g ClusterEC.gff -b ClusterEC.out --cdd_cog_file $CONCOCT/scgs/cdd_to_cog.tsv > ClusterEC.cogsThen we determine those regions of the contigs with core COGs on in single copy using the 982 predetermined E. coli core COGs:
$DESMAN/scripts/SelectContigsPos.pl $DESMAN/complete_example/EColi_core_ident95.txt < ClusterEC.cogs > ClusterEC_core.cogsNow we just reformat the location of core cogs on contigs:
cut -d"," -f2,3,4 ClusterEC_core.cogs | tr "," "\t" > ClusterEC_core_cogs.tsv##Determine variants on core COGs
To input into bam-readcount:
cd ..
mkdir Counts
Before doing so though we need to index the contigs fasta file
samtools faidx contigs/final_contigs_c10K.fathen run bam-readcount:
for file in Map/*sorted.bam
do
stub=${file%.mapped.sorted.bam}
stub=${stub#Map\/}
echo $stub
(bam-readcount -q 20 -l AnnotateEC/ClusterEC_core_cogs.tsv -f contigs/final_contigs_c10K.fa $file 2> Counts/${stub}.err > Counts/${stub}.cnt)&
doneNext we collate the positions frequencies into a single file for Desman, here we use all genes regardless of length:
$DESMAN/scripts/ExtractCountFreqP.pl AnnotateEC/ClusterEC_core.cogs Counts 0 > Cluster_esc3_scgs.freqNow lets use Desman to find the variant positions on these core cogs:
mkdir Variants
cd Variants/
mv ../Cluster_esc3_scgs.freq .
python $DESMAN/desman/Variant_Filter.py Cluster_esc3_scgs.freq
cd ..and run Desman:
mkdir RunDesman
cd RunDesman
for g in 2 3 4 5 6 7 8; do
for r in 0 1 2 3 4; do
desman ../Variants/outputsel_var.csv -e ../Variants/outputtran_df.csv -o ClusterEC_${g}_${r} -r 1000 -i 100 -g $g -s $r > ClusterEC_${g}_${r}.out&
done;
done
cd ..First lets have a look at the mean posterior deviance as a function of strain number:
cat */fit.txt | cut -d"," -f2- > Dev.csv
sed -i '1iH,G,LP,Dev' Dev.csv which we can plot with a simple R script included in the Desman distribution:
cd $DESMAN_EXAMPLE
$DESMAN/scripts/PlotDev.R -l RunDesman/Dev.csv -o RunDesman/Dev.pdfFrom this it is not as clear as in the full data set analysed in the paper that five strains are present, since on average there is some improvement going from five to six strains. However, one particular run with five strains is as good as the runs with six, we should prefer the run with best fit for smallest strain number, so we shall use this.
To validate the strain inference we will download pre-identified sequences for each of the 982 single copy core COGs in the five known reference genomes.
cd $DESMAN_EXAMPLE
mkdir Validate
cd Validate
wget https://www.dropbox.com/s/f6ojp1qt4fz5lzn/Hits.tar.gz
tar -xvzf Hits.tar.gz
We then select core COGs that were included in our analysis. We reverse those that are reversed on the contigs so that positions match and then find all variants mapping onto the 0,1 encoding employed in DESMAN.
mkdir Select
$DESMAN/scripts/Select.sh
$DESMAN/scripts/ReverseStrand.pl ../AnnotateEC/ClusterEC_core.cogs
$DESMAN/scripts/TauFasta.pl
$DESMAN/scripts/CombineTau.pl > ClusterEC_core_tau.csv
We then compare these known assignments to those predicted by DESMAN:
python $DESMAN/scripts/validateSNP.py ../RunDesman/ClusterEC_5_0/Collated_Tau_mean.csv ClusterEC_core_tau.csv
The output should look like:
[[ 0.58724428 0.28880866 0.58935018 0.37184116 0.03610108]
[ 0.06257521 0.59596871 0.32190132 0.60830325 0.5631769 ]
[ 0.34115523 0.59055355 0.08092659 0.60469314 0.55054152]
[ 0.60409146 0.40222623 0.61101083 0.03188929 0.37304452]
[ 0.60108303 0.03820698 0.60018051 0.4076414 0.28399519]]
This gives for each (row) predicted haplotype (or posterior mean in fact) the fraction of SNPs differing to each of the five reference genomes (columns). We see that each strain maps to one genome with an error rate between 3.2% and 8.1%.
##Determine accessory genomes
Now we need the variant frequencies on all contigs:
cd $DESMAN_EXAMPLE
$DESMAN/scripts/Lengths.py -i AnnotateEC/ClusterEC.fa > AnnotateEC/ClusterEC.len
mkdir CountsAll
$DESMAN/scripts/AddLengths.pl < AnnotateEC/ClusterEC.len > AnnotateEC/ClusterEC.tsv
for file in Map/*sorted.bam
do
stub=${file%.mapped.sorted.bam}
stub=${stub#Map\/}
echo $stub
(bam-readcount -w 1 -q 20 -l AnnotateEC/ClusterEC.tsv -f contigs/final_contigs_c10K.fa $file > CountsAll/${stub}.cnt 2> CountsAll/${stub}.err)&
doneWe also need to extract info on all genes in the E. coli clusters:
python $DESMAN/scripts/ExtractGenes.py -g AnnotateEC/ClusterEC.gff > AnnotateEC/ClusterEC.genes
Then we collate the count files together filtering to genes greater than 500bp:
$DESMAN/scripts/ExtractCountFreqP.pl AnnotateEC/ClusterEC.genes CountsAll 500 > Cluster_esc3.freq
and find variants this time insisting on a minimum frequency of 3% and not filtering on sample coverage:
mkdir VariantsAll
cd VariantsAll
mv ../Cluster_esc3.freq .
python $DESMAN/desman/Variant_Filter.py Cluster_esc3.freq -m 0.0 -v 0.03
cd ..
To assign contigs we also need individual gene coverages, for consistency we generate these from the aggregated count files:
cd VariantsAll
python $DESMAN/scripts/CalcGeneCov.py Cluster_esc3.freq > Cluster_esc3_gene_cov.csv
Get list of core COGs:
cut -d"," -f5 ../AnnotateEC/ClusterEC_core.cogs > ClusterEC_core_genes.txt
Calculate coverage on core genes:
python $DESMAN/scripts/CalcDelta.py Cluster_esc3_gene_cov.csv ClusterEC_core_genes.txt ClusterEC_core
Select run with lowest deviance and 5 strains:
export SEL_RUN=$DESMAN_EXAMPLE/RunDesman/ClusterEC_5_0/
Then we run the gene/contig assignment algorithm.
python $DESMAN/desman/GeneAssign.py ClusterEC_coremean_sd_df.csv $SEL_RUN/Gamma_star.csv Cluster_esc3_gene_cov.csv $SEL_RUN/Eta_star.csv -m 20 -v outputsel_var.csv -o ClusterEC --assign_tau > ClusterEC.cout&
This should generate the following output files.
-
ClusterEC_log_file.txt: A log file
-
ClusterECeta_df.csv: The assignments from NMF unmanipulated useful for identifying multicopy genes.
-
ClusterECetaD_df.csv: As above but discretised NMF predictions.
-
ClusterECetaS_df.csv: Predictions from the Gibbs sampler selecting run with maximum log posterior.
-
ClusterECetaM_df.csv: Mean log posterior predictions from Gibbs sampler.
##Validate accessory genomes
We will now compare predictions with known assignments to reference genomes. First we use the mapping files to determine number of reads from each genome mapping to each gene.
python $DESMAN/scripts/gene_read_count_per_genome.py ../contigs/final_contigs_c10K.fa ../AnnotateEC/ClusterEC.genes ../AssignGenome/Mock1_20genomes.fasta ../Map/*mapped.sorted.bam > ClusterEC_gene_counts.tsv
As above we will rename the header file to be a bit more presentable:
$DESMAN/scripts/MapGHeader.pl $DESMAN/complete_example/Map.txt < ClusterEC_gene_counts.tsv > ClusterEC_gene_countsR.tsv
and select just unambiguous assignments to E. coli genomes:
cut -f1-6 < ClusterEC_gene_countsR.tsv > ClusterEC_gene_counts_unamb.tsv
We then do a little bit of R to convert the counts into gene assignments to genomes assuming that if more than 1% of reads mapping to a gene derive from a genome then that gene is present in that genome.
R
Gene_eta <- read.table("ClusterEC_gene_counts_unamb.tsv",header=TRUE,row.names=1)
Gene_etaP <- Gene_eta/rowSums(Gene_eta)
Gene_etaP[Gene_etaP > 0.01] = 1.
Gene_etaP[Gene_etaP <= 0.01] = 0.
write.csv(Gene_etaP,"Gene_etaP.csv",quote=FALSE)
Final we compare the mean posterior predictions to those assignments.
python $DESMAN/scripts/CompAssign.py ClusterECetaM_df.csv Gene_etaP.csv
Output should look like:
0.9791
0.9624
0.9663
0.9431
0.9657
Av. accurracy = 0.963333