This is an archived version of the 3D filament annotator.
For an up-to-date version, see here
Annotation of filament / curvilinear structures in 3D based on napari viewer.
conda create -n myenv python=3.9.0
conda activate myenv
pip install napari[all]
pip install git+https://github.com/amedyukhina/filament_annotator_3D.git
from skimage import io
image = io.imread('img/example_image.tif')import napari
viewer = napari.view_image(image, ndisplay=3)from filament_annotation_3d.annotator import add_annotation_layer, annotate_filaments
an_layer = add_annotation_layer(viewer)
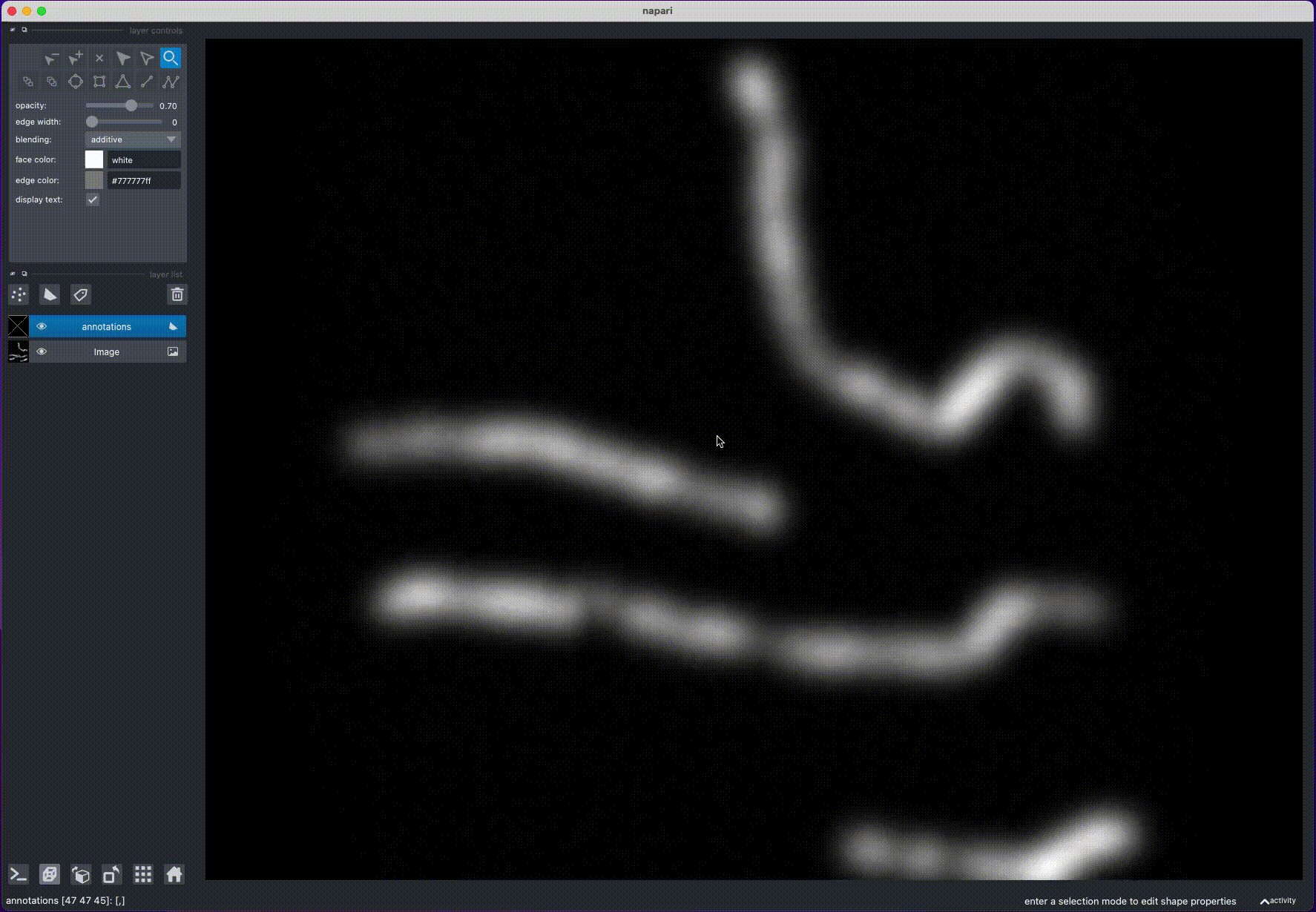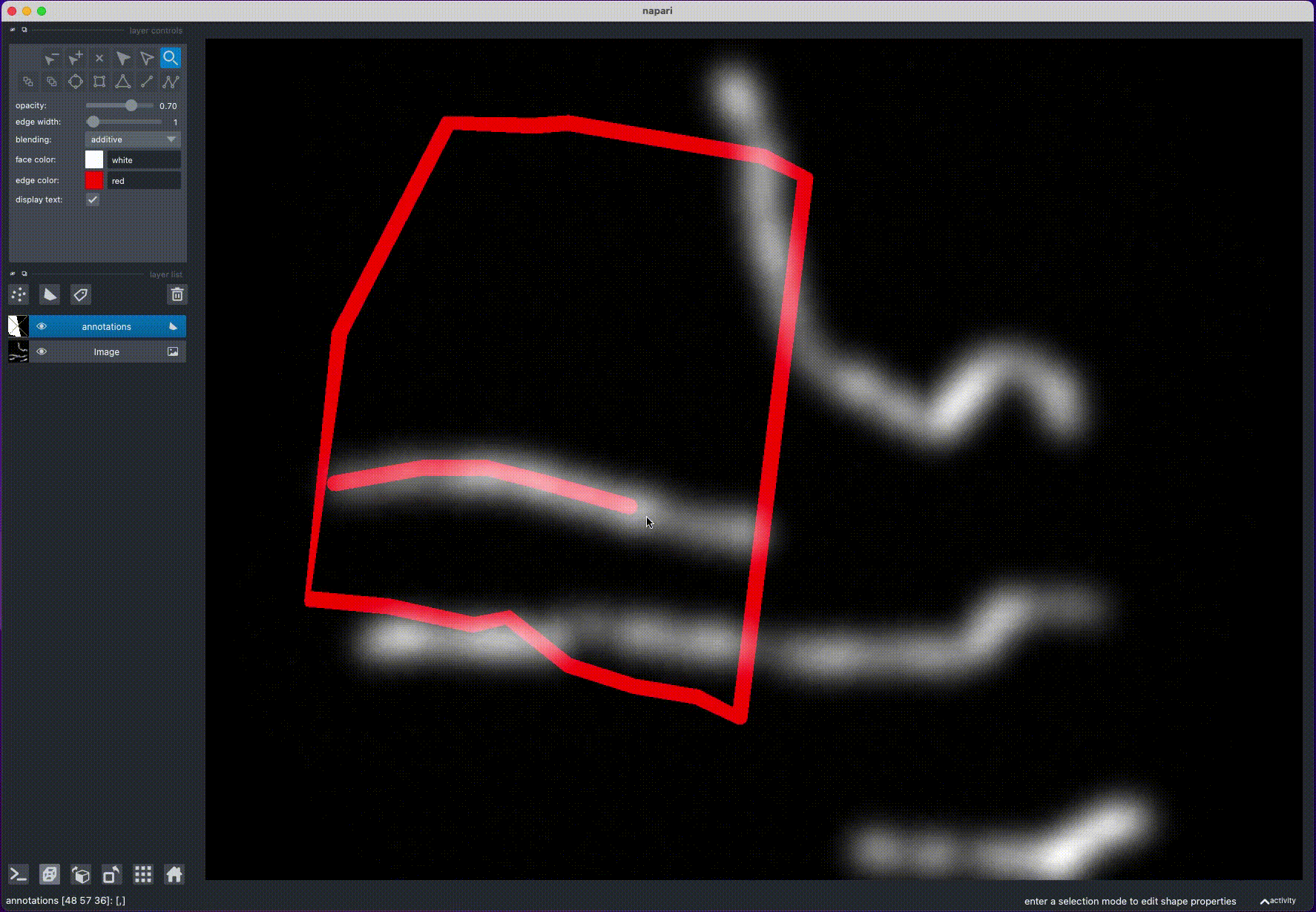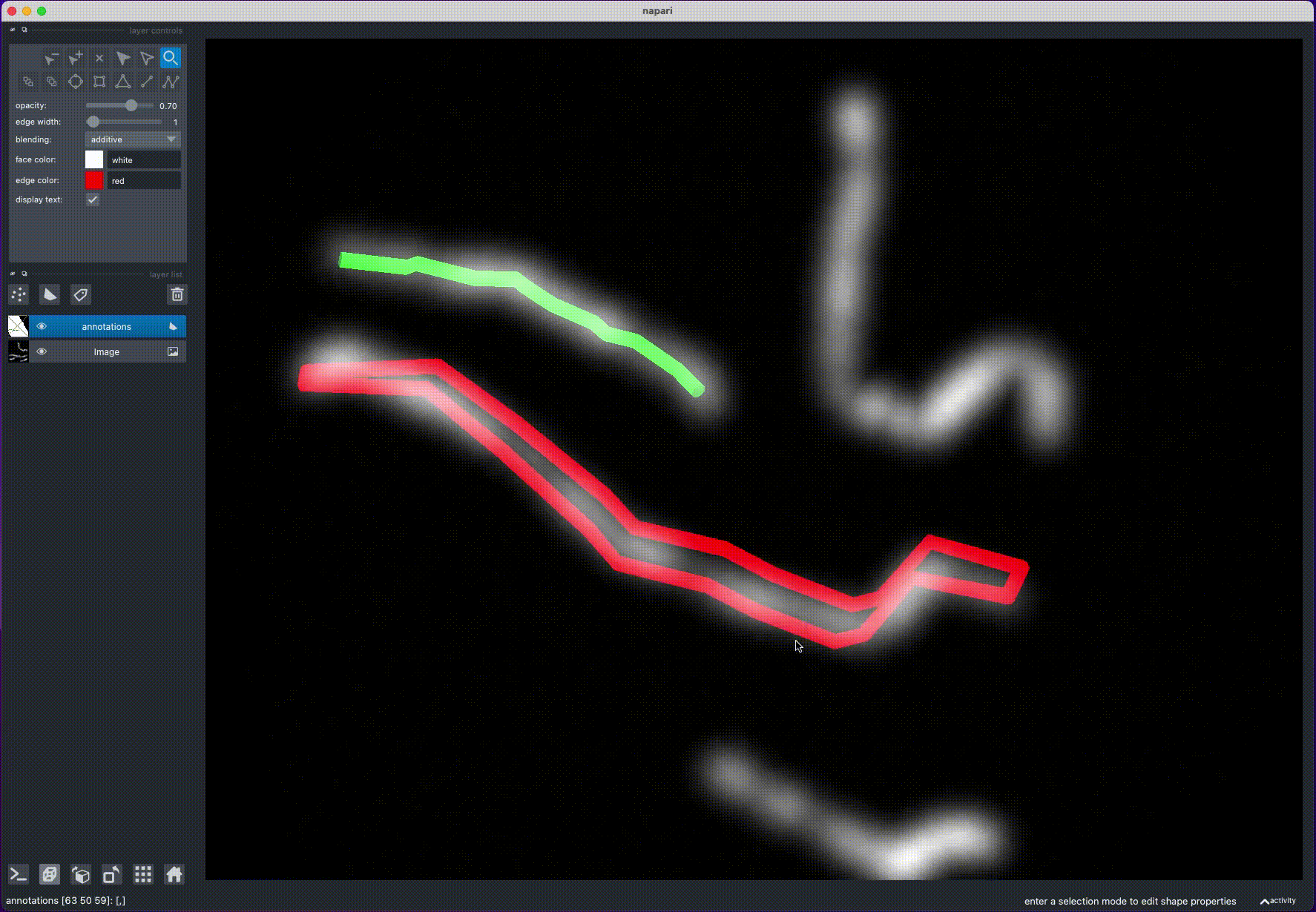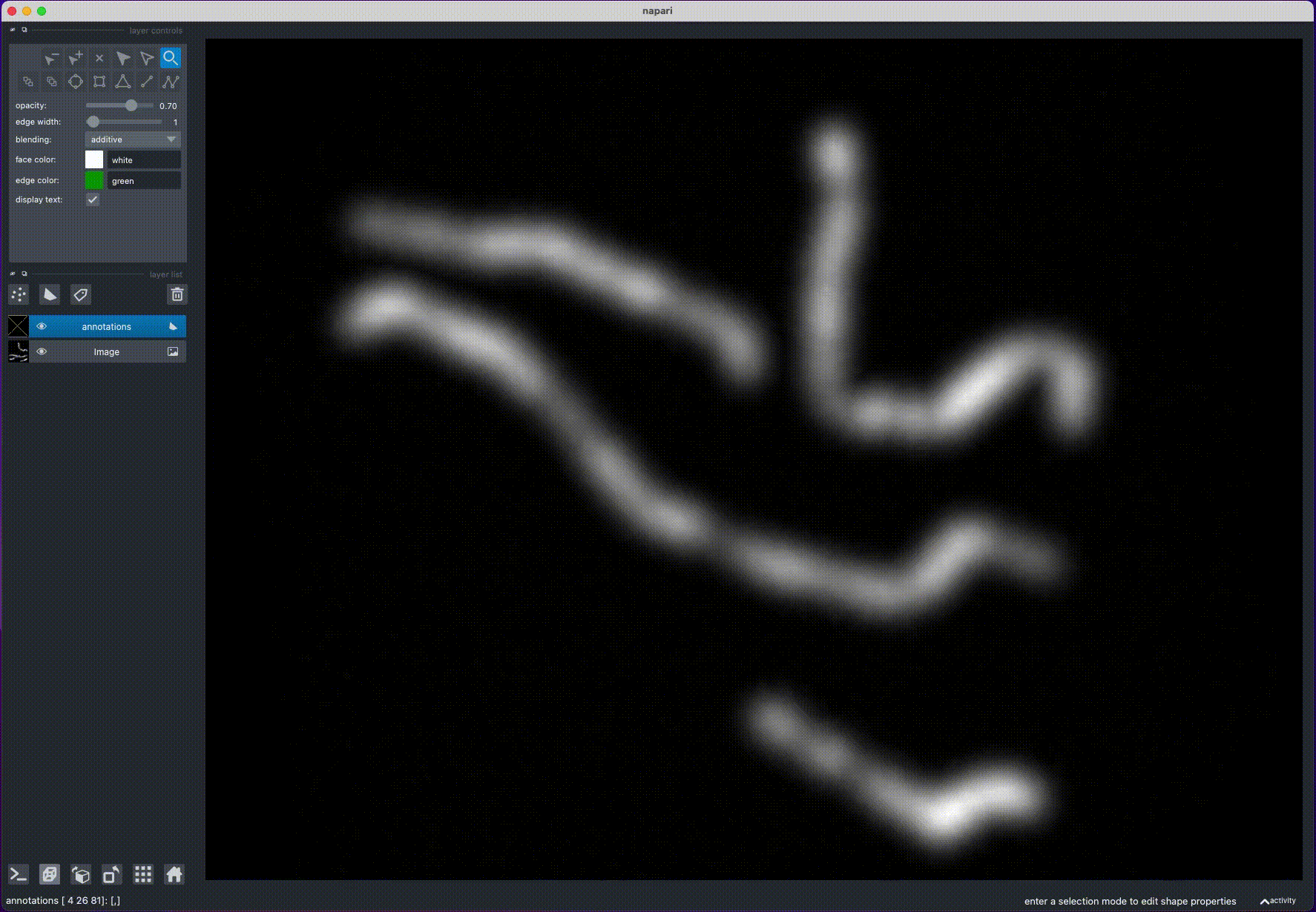
annotate_filaments(an_layer, 'annotations.csv') - Rotate the image to find a position, where the filament is clearly visible
- Draw a line over the filament, by holding "Control" (or "Command" on MacOS) and clicking with the mouse: this will draw a polygon with potential filament locations
- Rotate the image to view the filament from another angle and repeat step 2
- Rotate the image again: this will calculate the filament position from the intersection of the two polygons
- Repeat steps 1-4 for other filaments
To delete the last added point, press "d"
To delete the last added shape (polygon or filament), press "p"
The annotation results will be automatically saved in the csv file,
provided when running annotate_filaments.
To account for different pixel size in z and xy, provide the scale parameter
when starting the napari viewer, as a list of values for [z, y, x]:
viewer = napari.view_image(image, ndisplay=3, scale=[0.3, 0.1, 0.1]) This will automatically set the proper scale when creating the annotation layer. The output annotations will be saved in the original image scale (pixels).
The thickness of the lines is specified by the point_size parameter
of the annotate_filaments function (default value is 1):
annotate_filaments(an_layer, 'annotations.csv', point_size=0.5)There is an option to "snap" the annotated points to the brightest point in the neighborhood by specifying an image layer to use for snapping:
annotate_filaments(an_layer, 'annotations.csv', image_layer=viewer.layers[0]) The "snapping" can be fine-tuned by adjusting the following parameters:
sigma: Gaussian sigma (in pixels) used to smooth the image layer.
Can be provided as a list of value for each dimension. Default value is 2.
neighborhood_radius: Radius of the neighborhood (in pixels)
to consider for identifying the brightest points.
Can be provided as a list of value for each dimension. Default value is 5.
decay_sigma: Gaussian sigma (in pixels) used to scale image intensities
in the neighborhood patch to give preference to the originally annotated point.
The neighborhood patch centered on the originally annotated point
is multiplied by a Gaussian kernel with the provided sigma value
to scale down the pixels that are further away from the original point.
This is done to prevent "snapping" to another filament
located in the neighborhood.
The value of decay_sigma can be provided as a list of value for each dimension.
Default value is 5.
The optimal values for these parameters will depend on the level of noise, the thickness of the filaments and how close they are to each other.
Usage example:
annotate_filaments(an_layer, 'annotations.csv',
image_layer=viewer.layers[0],
sigma=[0.5, 1.5, 1.5],
neighborhood_radius=[2, 6, 6],
decay_sigma=[1, 3, 3])