This repository allows to reproduce the results in Neural Fine Gray - Please use the release: CHIL for exact reproduction.
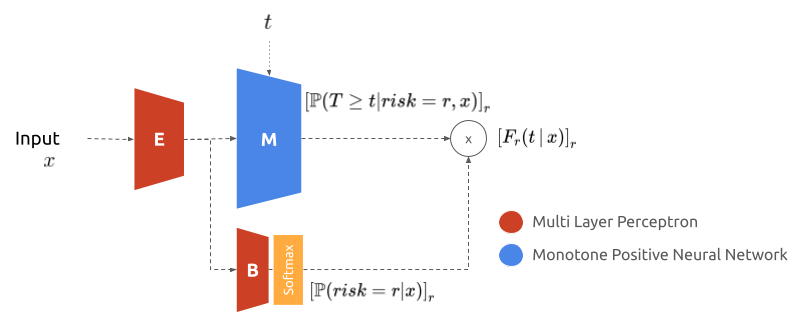
A neural network approach to the problem of competing risks, leveraging monotone neural networks to model the cumulative incidence functions.
The model consists in two neural networks: one models the cumulative incidence function and the other the balance to ensure that they add up to one.
To use the model, one needs to execute:
from nfg import NeuralFineGray
model = NeuralFineGray()
model.fit(x, t, e)
model.predict_risk(x, risk = 1)With x, the covariates, t, the event times and e, the cause of end of follow up (0 is censoring).
A full example with analysis is provided in examples/Neural Fine Gray on FRAMINGHAM Dataset.ipynb.
To reproduce the paper's results:
- Clone the repository with dependencies:
git clone git@github.com:Jeanselme/NeuralFineGray.git --recursive - Create a conda environment with all necessary libraries
pycox,lifelines,pysurvival - Add path
export PYTHONPATH="$PWD:$PWD/DeepSurvivalMachines:$PYTHONPATH" - Run
examples/experiment_competing_risk.py FRAMINGHAMto run all models on theFRAMINGHAMdataset - Repeat with
PBC,SYNTHETIC_COMPETINGandSEERto run on each dataset - Analysis using
examples/Analysis.ipynbto measure performance
Note that you will need to export the SEER dataset from https://seer.cancer.gov/data/. The previous scripts allow you to reproduce all the models presented in the paper except the Fine-Gray appraoch that requires:
0. Install R and the libraries: riskRegression, prodlim, survival, cmprsk and readr
- Create a folder
data/inexamples/to save the generated files - Run
examples/process_data.py FRAMINGHAMto create a csv files with the same data split used in the Python scripts - Run
examples/FineGray.Rto create the predictions of a Fine-Gray model (Note that you will need to change the content of this file for running on a subset of datasets)
Adding a new method consists in adding a child to Experiment in experiment.py with functions to compute the nll and fit the model.
Then, add the method in examples/experiment_competing_risk.py and follow the previous point.
TODOs have been added to make the addition of a new method easier.
We followed the same architecture than the DeepSurvivalMachines repository with the model in nfg/ - only the api should be used to test the model. Examples are provided in examples/.
git clone git@github.com:Jeanselme/NeuralFineGray.git --recursive
The model relies on DeepSurvivalMachines, pytorch, numpy and tqdm.
To run the set of experiments pycox, lifelines, pysurvival are necessary.