Paper PDF
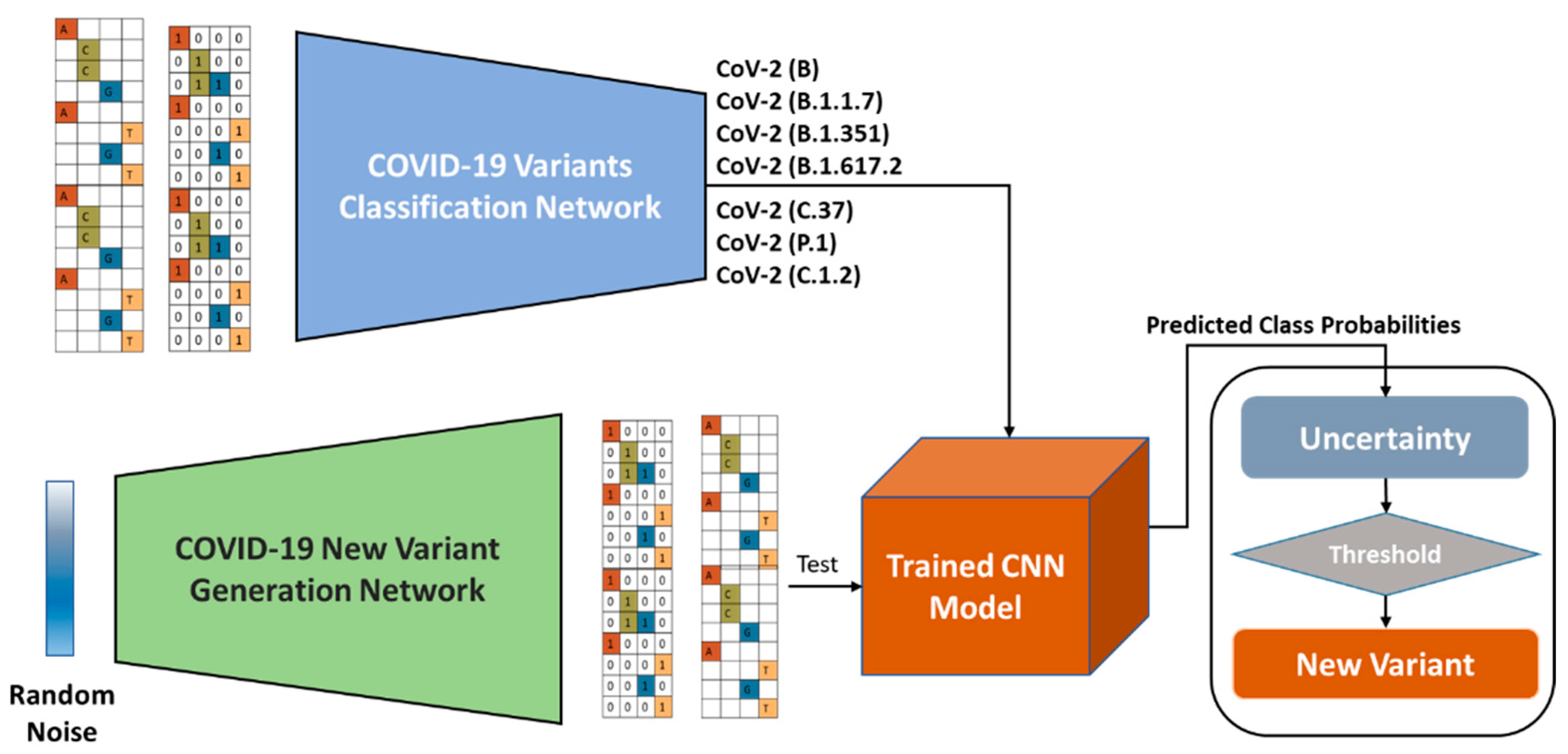
This code is an implementation of a deep learning model for classifying COVID-19 variants based on their nucleotide or protein sequence. The model architecture is defined in MyModels.py file and Conv1DModel() function is used to define the model. The model is trained using the train_test_split function from the sklearn.model_selection module and the dataset is loaded from Utils.py file.
#Requirements The following packages need to be installed to run this code:
numpy tensorflow matplotlib scikit-learn
The dataset used for training and testing the model consists of nucleotide or protein sequences of various COVID-19 variants. The sequences are stored in the data/genomic.fna and data/protein.faa files respectively. The data can be downloaded from NCBI website https://www.ncbi.nlm.nih.gov/ with the following keywords in the search:
CoV-2 (B) CoV-2 (B.1.1.7) CoV-2 (B.1.351) CoV-2 (B.1.617.2) CoV-2 (C.37) CoV-2 (P.1) CoV-2 (B.1.525)
The sequences of other COVID-19 variants can be used for testing the model by uncommenting the code that loads the data for other variants in the Utils.py file.
Usage To use this code, simply run the TrainCNN_COVID19Variants.py file. The code will train the model, plot the accuracy and loss curves, plot the ROC curve and confusion matrix and save the model in the current directory.