- Autoclave vials with lids on.
- For “open” vials, it is not necessary to autoclave the needles.
- Clean lines with sterile water (20s), and 70% ethanol (20s).
- Weigh empty vials, note OD of (optional: washed) dense cultures.
- Fill each vial with 25mL of media. Connect and run efflux lines with cleaned efflux needles. Weigh final culture volumes.
- Once vials are connected to the fluid lines set up the experiment.
Setting up the experiment:
- Open up an
iterm2terminal (Command+space iterm2). - We use
bashinstead of the default terminal. Type inbashand hit return.- If this was done right, the prompt should change, and should be preceded by something like
(py37)which indicates that the virtual environment has been loaded.
- If this was done right, the prompt should change, and should be preceded by something like
- In the file browser (“finder”), navigate to
Documents/dpu/experiments/. - Duplicate the
evolver/folder, and name the duplicate with a descriptive name. TypicallyYYYY_MM_DD_keywords_separted_by_underscore. - Switch back to the terminal window and type in
cd ~/Documents/dpu/experiment/NAME_OF_YOUR_NEW_DIRECTORY - Start the visual interface to populate the configuration file by
typing in
streamlit run visualize.py. This will return a localhost IP address with a port. Copy and paste this in a browser window. - Populate all the fields, and press ‘Write to configuration file’. NOTE: If you are running a calibration, or if you are not planning on using a calibration file, LEAVE THE “Calibration name” field BLANK!!!
- Alternative approach: The folder has an
example_experiment_parameters.yamlfile. Edit the contents of this file and save it to a file calledexperiment_parameters.yamlin the same directory. - If you are running a calibration:
- Include the word “-calibration” in the experiment name.
- Currently, if the per vial parameter
calib_end_odis left at its default values (0), it is assigned a valuecalib_start_od/10. Please explicitly specify this parameter taking into consideration the sensor readings saturate around 0.075. - Open a new terminal window, load bash, navigate to the
experiment folder, and start the calibration by typing in
python eVOLVER.py. This will display the calibration settings. When prompted about blanking vials, type n. - The calibration will stop at the last step. Collect the efflux from each tube. Select all the vials from the Rpi touch interface on the eVOLVER and run the efflux for 10s. Measure the ODs. Multiply the ODs with a factor of 1.2 if measuring efflux culture.
- Go back to the web interface, update the
calib_end_odwith the new ODs, write this to file again. NB: The old file will be automatically renamed and time stamped. - Open a new terminal, load bash, navigate to the experiment directory as in STEP 5.
- Type in
python write_calibration_to_file.py. This will create a CSV file with the name of the calibration experiment, the same as the experiment name. - Next, switch back to the browser and type in the name of this calibration. The real experiment will need to have a different name.
- When the experiment config file is filled, switch to the terminal
and in a new tab (load bash and navigate to the experiment
direcotory), type in
python eVOLVER.py - Sanity check the experiment settings, press
nwhen prompted for blanking the vials, and the experiment should start. - When the experiment is running, a new drop down option should be added corresponding to the experiment name, displaying all the plots.
- The major caveat in passing message to the Rpi with the current
design is that the strings are all expected to be 16 values
separated by commas. This is currently handled by looping over
vialswhich has a list of 16 integers, and populating lists like volumes, rates, and periods by looking up indices from other data sources like user defined variables. This is pretty low level right now, and there is no real API to arbitrarily control sleeve behavior without resorting to hacks like creating custom log files for history dependent control of sleeves - There are low level (arduino-level) event logging requirements
which are going to be important to debug. This will impact things
like:
- Frequency of data logging
- Types of data that are logged systematically (pump events in chemostat mode etc)
I am not a fan of this design, where the custom script is loaded in order to first load parameters, but control is then handed back to eVOLVER.py. Custom imports
- Ideally, the config file is read once, and parameters are passed around in some data class or similar. See Issue http:github.com/amoghpj/evolver/issues/1.
- No consistent API in adding custom experiment handling. Need to move away from using per-vial logic in a loop like it is currently setup.
- Define defaults for stirring control that are passed explicitly to each function definition
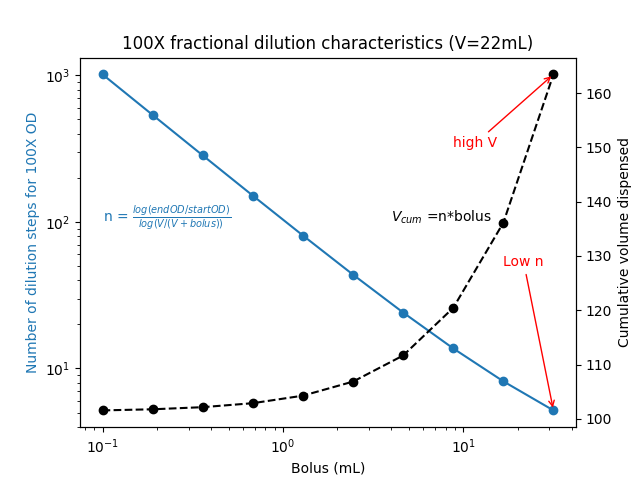
We can perform an in-vial calibration by diluting out a dense culture by dispensing media of fixed volumes repeatedly. Interestingly, performing a 100X dilution in steps uses far less media than doing it in a single step. The total volume of media consumed as the bolus size → 0 is - volume_vial * log(endOD/startOD).
Below is a visual depiction of the relationship between bolus size, number of steps needed to hit endOD, and the total volume dispensed.