IMPORTANT This project is currently is in development: please visit the legacy codebase: G2P2C
RL4T1D is a project to develop Reinforcement Learning (RL)-based Artificial Pancreas Systems (APS), with the aim to automate treatment in Type 1 Diabetes (T1D). For more detailed information please visit the legacy codebase: G2P2C
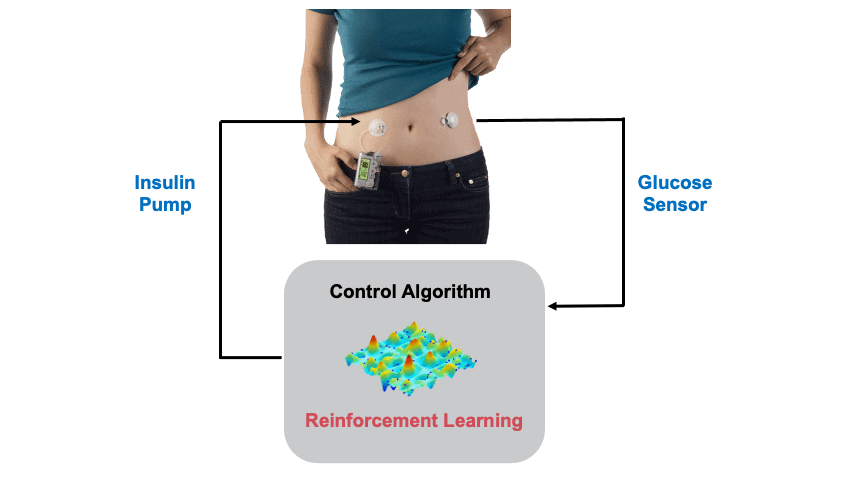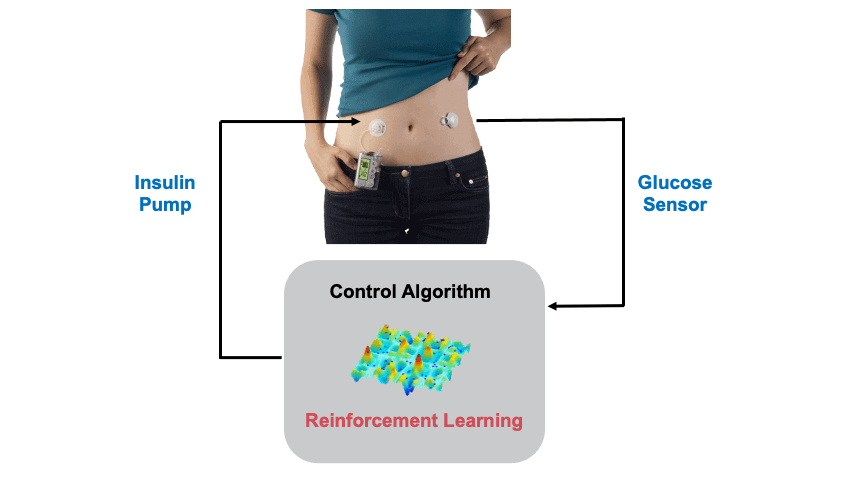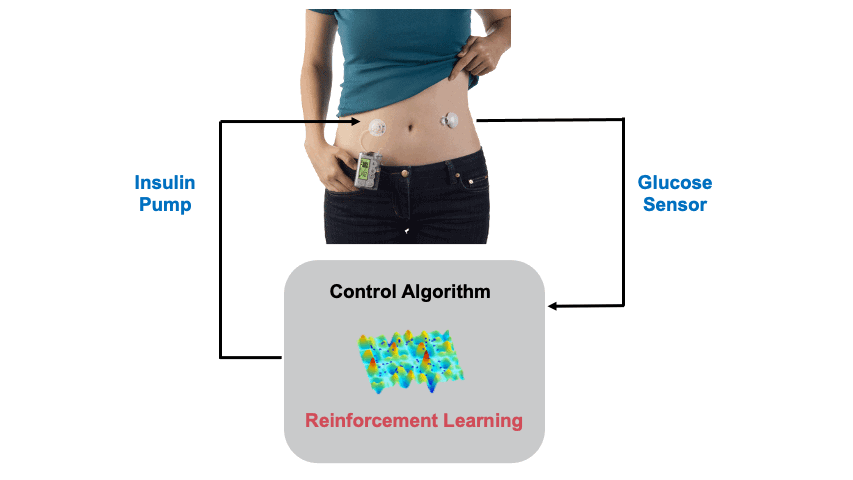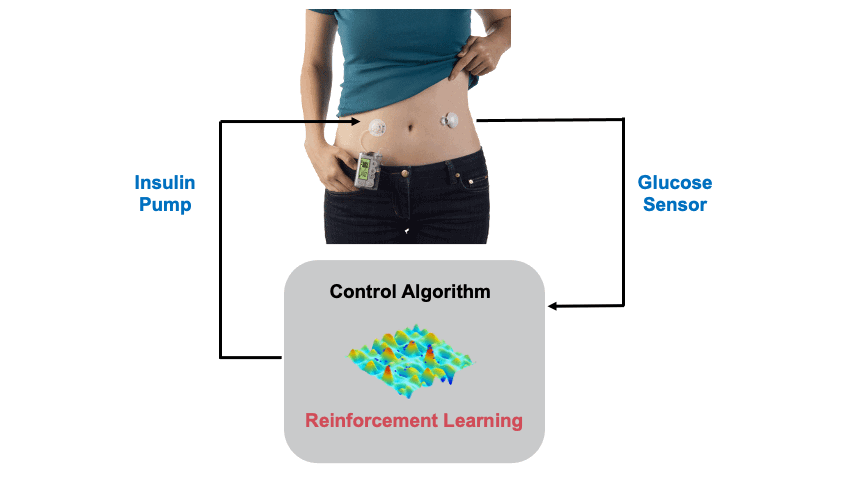
Background: Type 1 Diabetes (T1D) is casued by the autoimmune destruction of the islet beta-cells and results in absolute insulin deficiency (cover image: Human islet of Langerhans created by Stable Diffusion). Hence, external administration of insulin is required to maintain glucose levels, which is cruicial as both low and high glucose levels are detrimental to health. This is usually done through an insulin pump attached to the body. An continuous glucose sensor is also attached to measure the glucose levels so that a control algorithm can estimate the appropriate insulin dose. In this project we design Reinforcement Learning (RL) based Artificial Pancreas Systems (APS) for the glucose control problem. The figure below shows the main components of an APS.
Objective/Task: Maintaining glucose levels is a life-long optimisation problem, complicated due to the disturbances associated with daily events (meals, exercise, stress.. etc), delays present in glucose sensing and insulin action, partial observability, and safety constraints among others. A simulation of glucose regulation, using a RL-based strategy is shown below, where the optimal glucose range is shaded in green severe hypoglycemia / hyperglycemia ranges highlighted by the red dotted line. The blood glucose measurements are presented in the top, while the administered insulin by the RL agent is presented in the bottom. The disturbances related to meal events and the carbohydrate content of the meals are presented in red.
- Create and activate a Python3.10.12 virtual environment.
- Clone the repository:
git clone git@github.com:RL4H/RL4T1D.git. - Go to the project folder (RL4T1D):
cd RL4T1D - Install the required Python libraries
requirements.txt. - Create an environment file
.envat the root of the project folder withMAIN_PATH=path-to-this-project(echo "MAIN_PATH=$(pwd)">.env).
* You can also install from the source (Recommended to install using
pip install -e ., the simglucose 0.2.2 source code is available in the environments folder). The simulation environment and scenarios used in this project are extended from the original environment.
Running a Proximal Policy Optimisation (PPO) algorithm for glucose control. More information related to state-action space, reward formulations: Paper .
cd experiments
python run_RL_agent.py experiment.name=test1 experiment.device=cpu agent=ppo agent.debug=True hydra/job_logging=disabled
Running a Soft Actor Critic (SAC) algorithm for glucose control.
cd experiments
python run_RL_agent.py experiment.name=test2 experiment.device=cpu agent=sac agent.debug=True hydra/job_logging=disabled
Start mlflow
mlflow ui --backend-store-uri results/mlflow
A simple example is provided in simple_example.py.
# Setup MAIN_PATH.
from environment.t1denv import T1DEnv
from agents.algorithm.ppo import PPO
from utils.control_space import ControlSpace
from utils.logger import Logger
from utils.load_args import load_arguments
args = load_arguments(overrides=["experiment.name=test2", "agent=ppo", "env.patient_id=0", "agent.debug=True",
"hydra/job_logging=disabled"])
# print(vars(args))
agent = PPO(args=args.agent, env_args=args.env, logger=Logger(args), load_model=False, actor_path='', critic_path='')
env = T1DEnv(args=args.env, mode='testing', worker_id=1)
# to improve the training efficiency, we use a non-linear action space.
controlspace = ControlSpace(control_space_type=args.agent.control_space_type, insulin_min=env.action_space.low[0],
insulin_max=env.action_space.high[0])
observation = env.reset()
for _ in range(10):
rl_action = agent.policy.get_action(observation) # get RL action
pump_action = controlspace.map(agent_action=rl_action['action'][0]) # map RL action => control space (pump)
observation, reward, is_done, info = env.step(pump_action) # take an env step
print(f'Latest glucose level: {info["cgm"].CGM:.2f} mg/dL, administered insulin: {pump_action:.2f} U.')
# OR simply use agent.run() to train the agent.
print('Done')Running a clinical treatment algorithm: Basal-Bolus (BB). The parameters if the BB treatment can be updated via clinical_treatment.yaml file. More info: Paper .
cd experiments
python run_clinical_treatment.py experiment.name=test3 agent=clinical_treatment hydra/job_logging=disabled
A simple example is provided in simple_example.py.
# Setup MAIN_PATH.
from environment.t1denv import T1DEnv
from clinical.basal_bolus_treatment import BasalBolusController
from clinical.carb_counting import carb_estimate
from utils.load_args import load_arguments
from environment.utils import get_env, get_patient_env
from utils import core
args = load_arguments(overrides=["experiment.name=test2", "agent=clinical_treatment", "env.patient_id=0", "agent.debug=True", "hydra/job_logging=disabled"])
patients, env_ids = get_patient_env()
env = T1DEnv(args=args.env, mode='testing', worker_id=1)
clinical_agent = BasalBolusController(args.agent, patient_name=patients[args.env.patient_id], sampling_rate=env.env.sampling_time)
observation = env.reset() # observation is the state-space (features x history) of the RL algorithm.
glucose, meal = core.inverse_linear_scaling(y=observation[-1][0], x_min=args.env.glucose_min, x_max=args.env.glucose_max), 0
# clinical algorithms uses the glucose value, rather than the observation-space of RL algorithms, which is normalised for training stability.
for _ in range(10):
action = clinical_agent.get_action(meal=meal, glucose=glucose) # insulin action of BB treatment
observation, reward, is_done, info = env.step(action[0]) # take an env step
# clinical algorithms require "manual meal announcement and carbohydrate estimation."
if args.env.t_meal == 0: # no meal announcement: take action for the meal as it happens.
meal = info['meal'] * info['sample_time']
elif args.env.t_meal == info['remaining_time_to_meal']: # meal announcement: take action "t_meal" minutes before the actual meal.
meal = info['future_carb']
else:
meal = 0
if meal != 0: # simulate the human carbohydrate estimation error or ideal scenario.
meal = carb_estimate(meal, info['day_hour'], patients[id], type=args.agent.carb_estimation_method)
glucose = info['cgm'].CGM
print(f'Latest glucose level: {info["cgm"].CGM:.2f} mg/dL, administered insulin: {action[0]:.2f} U.')
print('Done')- Now you can use mlflow to organise: <experiments, experiment_runs (e.g., for hyperparams, new ideas)>.
- We have legacy custom visualisation scripts used to debug experiments while running - integrate with new flow
- Think/Complete logging. Duplicated with mlflow logs and legacy custom logging patterns
- validation results.
- Write test cases
- Add read docs like GluCoEnv.
- Improve handling offpolicy vs onpolicy workers.
- Issue running hydra from jupyter notebook.
- build batch scripts to run for mutiple seeds, envs.
- provide capability to run agents without using main script (integrate with other rl-libraries).
@misc{rl4t1d,
author={RL4H Team},
title={RL4T1D (2024)},
year = {2024},
publisher = {GitHub},
journal = {GitHub repository},
howpublished = {\url{https://github.com/RL4H/RL4T1D}},
}
@article{hettiarachchi2023reinforcement,
title={Reinforcement Learning-based Artificial Pancreas Systems to Automate Treatment in Type 1 Diabetes},
author={Hettiarachchi, Chirath},
year={2023},
publisher={The Australian National University}
}
@article{hettiarachchi2024g2p2c,
title={G2P2C—A modular reinforcement learning algorithm for glucose control by glucose prediction and planning in Type 1 Diabetes},
author={Hettiarachchi, Chirath and Malagutti, Nicolo and Nolan, Christopher J and Suominen, Hanna and Daskalaki, Elena},
journal={Biomedical Signal Processing and Control},
volume={90},
pages={105839},
year={2024},
publisher={Elsevier}
}
Chirath Hettiarachchi - chirath.hettiarachchi@anu.edu.au
School of Computing, College of Engineering & Computer Science,
Australian National University.