The aim of BPRMeth is to extract higher order features associated with the shape of methylation profiles across a defined genomic region. Using these higher order features across promoter-proximal regions, BPRMeth provides a powerful machine learning predictor of gene expression. Check the vignette on how to use the package. Modelling details for the different models can be found online: http://rpubs.com/cakapourani.
The original implementation has now been enhanced in two important ways: we introduced a fast, variational inference approach which enables the quantification of Bayesian posterior confidence measures on the model, and we adapted the method to use several observation models, making it suitable for a diverse range of platforms including single-cell and bulk sequencing experiments and methylation arrays.
To get the latest development version from Github:
# install.packages("devtools")
devtools::install_github("andreaskapou/BPRMeth", build_vignettes = TRUE)Or install from the stable release version from Bioconductor
## try http:// if https:// URLs are not supported
if (!requireNamespace("BiocManager", quietly=TRUE))
install.packages("BiocManager")
BiocManager::install("BPRMeth")You can the check the vignette on how to use the package:
browseVignettes("BPRMeth")If you get the following error when installing the package:
clang: error: unsupported option '-fopenmp'
try the following:
brew install llvm
brew install boost
brew install homebrew/science/hdf5 --enable-cxx
mkdir -p ~/.R
vim ~/.R/Makevars
## Paste the following commands
# The following statements are required to use the clang4 binary
CC=/usr/local/clang4/bin/clang
CXX=/usr/local/clang4/bin/clang++
CXX11=/usr/local/clang4/bin/clang++
CXX14=/usr/local/clang4/bin/clang++
CXX17=/usr/local/clang4/bin/clang++
CXX1X=/usr/local/clang4/bin/clang++
LDFLAGS=-L/usr/local/clang4/lib
# End clang4 inclusion statementsThese commands will point R to the new version of clang.
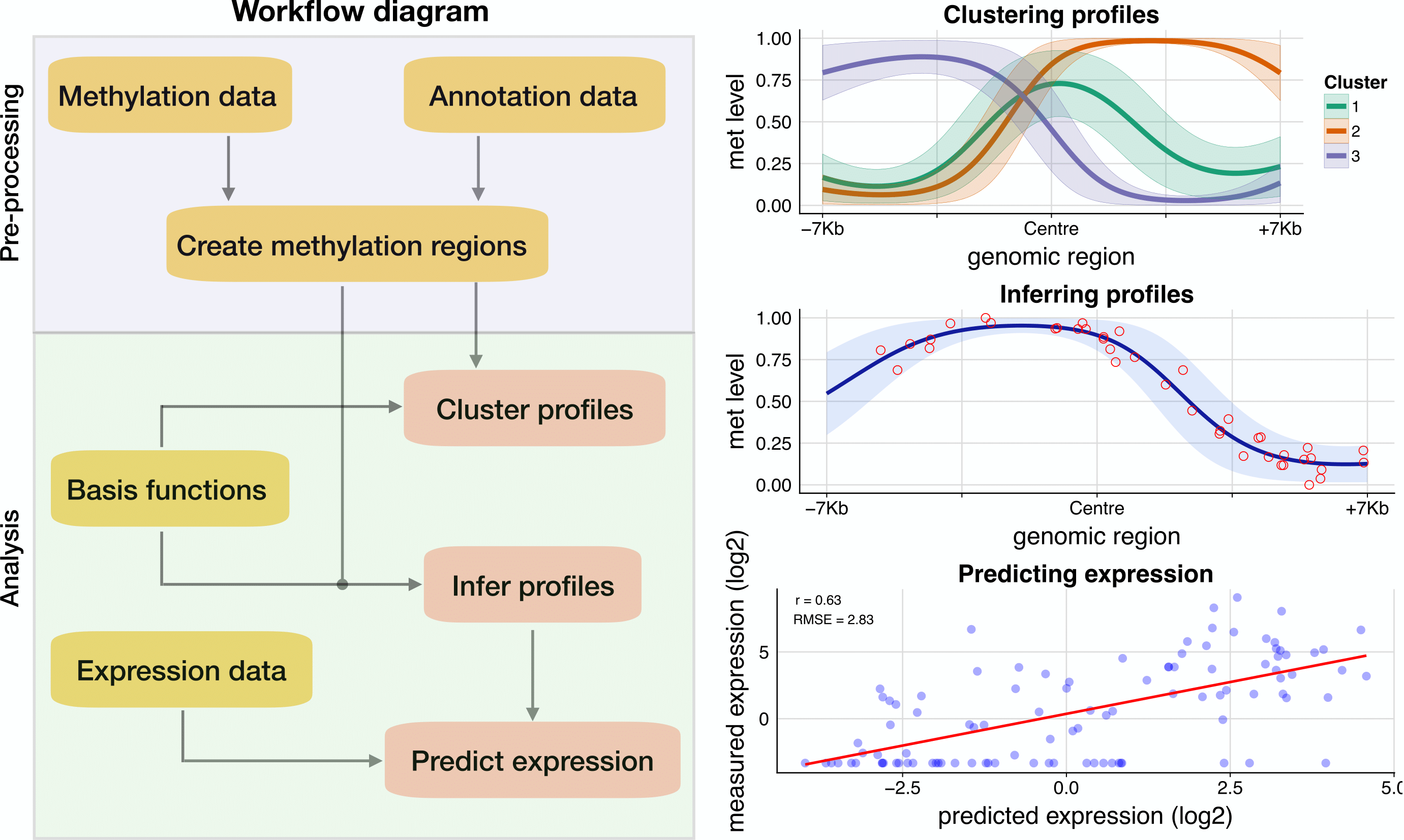
The diagram below shows an overview of the pre-processing and analysis workflow in BPRMeth, together with example output graphs.
Kapourani, C.-A. and Sanguinetti, G. (2016). Higher order methylation features for clustering and prediction in epigenomic studies. Bioinformatics 32 (17), i405-i412. (Best Paper Award in ECCB 2016).