Reference: BSc thesis Modeling the network dynamics underlying hippocampal sharp waves and sequence replay.
whole network retuned to match experimental parameters and moved to Brian2 (06.2017)
With the scripts in the repository one can create a CA3 network model and examine the network dynamics during hippocampal SWRs (Sharp Wave-Ripples).
To run the scripts, install Brian (version 1) and run:
git clone https://github.com/andrisecker/KOKISharpWaves.git # Clone this GitHub repository
cd KOKISharpWaves
mkdir figures # creates directory for the saved figures
cd scripts
# (on can found the generates spike trains in the files folder, or run the next line)
python generate_spike_train.py # generate CA3 like spike trains (as exploration of a maze) -> files/spikeTrainR.npz
python stdp_network_b.py # learns the recurrent weight (via STDP, based on the spiketrain) -> files/wmx.txt
python spw_network4a_1.py # creates the network, runs the simulation, extracts dynamic features
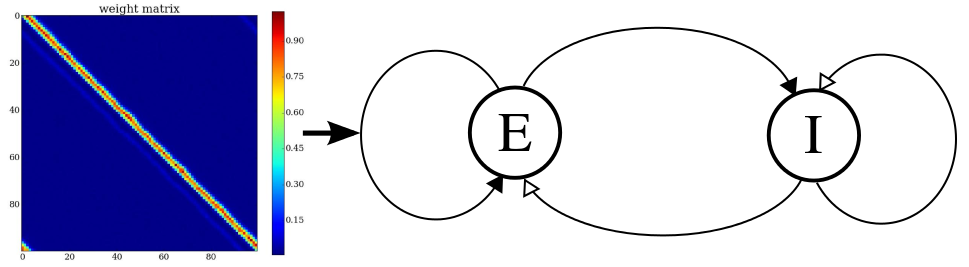
network structure with the learned recurrent weightmatrix
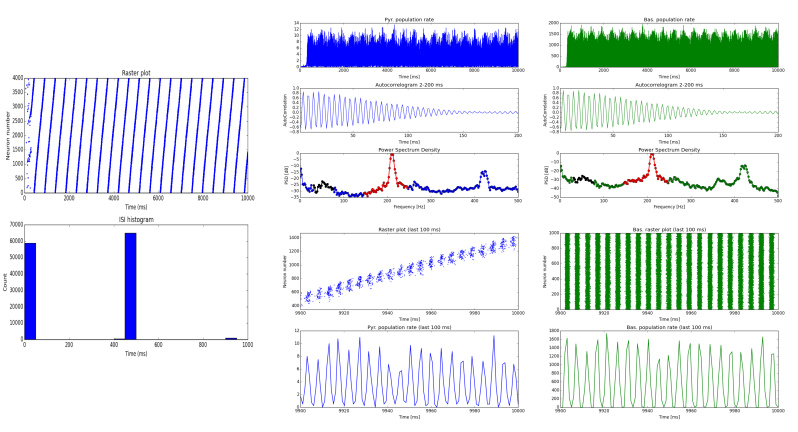
extraction of dynamic features during SWRs
python spw_network4_automatized.py # investigates into network dynamics with various scaling factor (of the weight matrix)
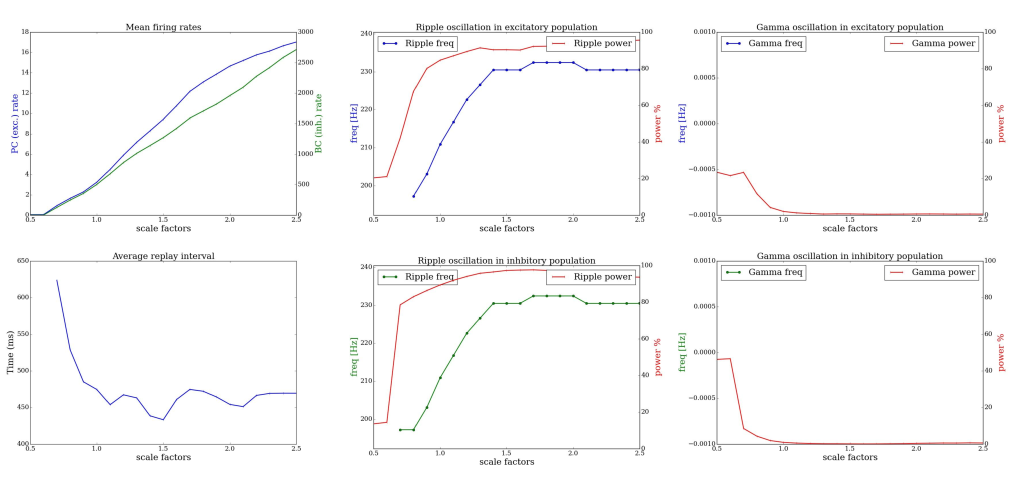
automated evaluation of the networks dynamics with differently scaled recurrent weightmatrix
python spw_network4_BasInputs_f.py # investigates into network dynamics with different outer inputs to the Basket cell population
python bayesian_inference.py # Bayesian decoding of place from spikes saved from spw...
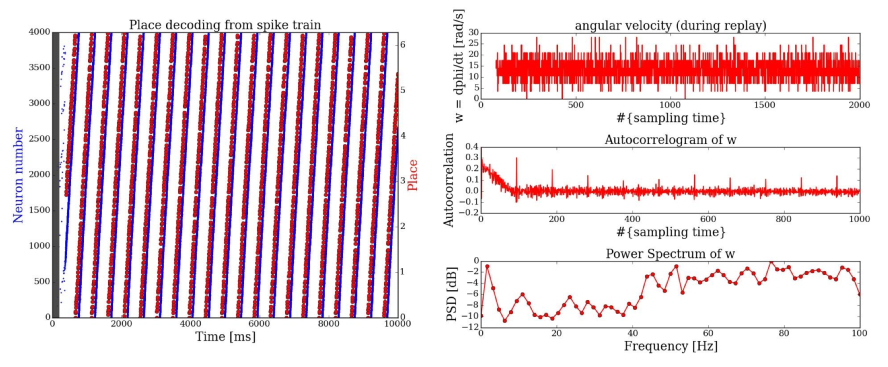
Bayesian decoding of place (in a circle maze) from spike trains recorded during SWRs
Raster plot of the activity and inferred place from spike trains + analysis of the angular velocity during SWRs
note: (with some IDE) one has to change the PATH (SWBasePath at the top of the scripts)