This is the development page of the stackr, if you want to help, see contributions section
Most genomic analysis look for patterns and trends with various statistics. Bias, noise and outliers can have bounded influence on estimators and interfere with polymorphism discovery. Avoid bad data exploration and control the impact of filters on your downstream genetic analysis. Use stackr to: import, explore, manipulate, visualize, filter, impute and export your GBS/RADseq data.
To try out the dev version of stackr, copy/paste the code below:
if (!require("devtools")) install.packages("devtools") # to install
devtools::install_github("thierrygosselin/stackr", build_vignettes = TRUE) # to install WITH vignettes
library(stackr) # to load| Caracteristics | Description |
|---|---|
| Import | Various supported genomic file formats in tidy_genomic_format and/or their separate module:VCF (Danecek et al., 2011) PLINK tped/tfam (Purcell et al., 2007) genind (Jombart et al., 2010; Jombart and Ahmed, 2011) genlight (Jombart et al., 2010; Jombart and Ahmed, 2011), also in tidy_genlightstrataG gtypes (Archer et al., 2016) Genepop (Raymond and Rousset, 1995; Rousset, 2008), also in tidy_genepopSTACKS haplotype file (Catchen et al., 2011, 2013) hierfstat (Goudet, 2005), also in tidy_fstatDataframes of genotypes in wide or long/tidy format, also in tidy_wide |
| Output | Export genomic data out of stackr using genomic_converter or the separate modules (12 formats):write_vcf: VCF (Danecek et al., 2011)write_plink: PLINK tped/tfam (Purcell et al., 2007)write_genind: adegenet genind and genlight (Jombart et al., 2010; Jombart and Ahmed, 2011)write_genlight: genlight (Jombart et al., 2010; Jombart and Ahmed, 2011)write_gtypes: strataG gtypeswrite_genepop: Genepop (Raymond and Rousset, 1995; Rousset, 2008)STACKS haplotype file (Catchen et al., 2011, 2013) write_betadiv: betadiv (Lamy, 2015)vcf2dadi: δaδi (Gutenkunst et al., 2009)write_structure: structure (Pritchard et al., 2000)write_arlequin: Arlequin (Excoffier et al. 2005)write_hierfstat: hierfstat (Goudet, 2005)Dataframes of genotypes in wide or long/tidy format |
| Conversion function | genomic_converter import/export genomic formats mentioned above. The function is also integrated with usefull filters, blacklist and whitelist. |
| Outliers detection | detect_duplicate_genomes: Detect and remove duplicate individuals from your dataset detect_mixed_genomes: Detect and remove potentially mixed individualssummary_haplotype and filter_snp_number: Discard of outlier markers with de novo assembly artifact (e.g. markers with an extreme number of SNP per haplotype or with irregular number of alleles) |
| Pattern of missingness | missing_visualization: Visualize patterns of missing data. Find patterns associated with different variables of your study (lanes, chips, sequencers, populations, sample sites, reads/samples, homozygosity, etc) |
| Filters | Alleles, genotypes, markers, individuals and populations can be filtered and/or selected in several ways:filter_coverage: Discard markers based on read depth (coverage) of alleles and genotypesfilter_genotype_likelihood: Discard markers based on genotype likelihoodfilter_individual: Discard markers based on proportion of genotyped individualsfilter_population: Discard markers based on proportion of genotyped populationsfilter_het: Discard markers based on the observed heterozygosity (Het obs)filter_fis: Discard markers based on the inbreeding coefficient (Fis)filter_hw: Discard markers based on the Hardy-Weinberg Equilibrium expectations (HWE)filter_maf: Discard markers based on local or global (overall) minor allele frequency |
| Imputations | Map-independent imputations of missing genotypes. Using Random Forest or the most frequent category. Imputations can be conducted overall samples or by populations. Imputations are integrated in several functions and in a separate module: stackr_imputations_module |
| ggplot2-based plotting | Visualize distribution of important metric and statistics and create publication-ready figures |
| Parallel | Codes designed and optimized for fast computations running imputations, iterations, etc. in parallel |
- Parallel computing: Follow the steps in this vignette to install an OpenMP enabled randomForestSRC package to do imputations in parallel.
- Installation problem: see this vignette
- Windows users: 1. Install Rtools. 2. To have stackr run in parallel, use parallelsugar. Easy to install and use (instructions).
- For a better experience in stackr and in R in general, I recommend using RStudio. The R GUI is unstable with functions using parallel (more info).
Below, the combination of packages and how I install/load them :
if (!require("pacman")) install.packages("pacman")
pacman::p_load(devtools, tidyverse, data.table, parallel, lazyeval, randomForestSRC)
devtools::install_github("thierrygosselin/stackr", build_vignettes = TRUE)
library("stackr")Vignettes (in development, check periodically for updates):
Vignettes can also be accessed inside R with:
browseVignettes("stackr") # To browse vignettes
vignette("vignette_vcf2dadi") # To open specific vignetteR Notebooks:
- Missing data visualization and analysis (html vignette) and (Rmd)
To get the citation, inside R:
citation("stackr")Change log, version, new features and bug history now lives in the NEWS.md file
v.0.4.5
-
temporary fix to
tidy_genomic_datato read unconventional Tassel VCF -
new function
ibdg_fhcomputes the FH measure that was previously computed insummary_haplotypes. It now works with biallelic and multiallelic data.The FH measure is based on the excess in the observed number of homozygous genotypes within an individual relative to the mean number of homozygous genotypes expected under random mating (see function for details). The
IBDgin the name is because the measure is a proxy of the realized proportion of the genome that is identical by descent by reference to the current population under hypothetical random mating. -
missing_visualizationnow computes the FH measure and look for correlation with average missingness per individual. -
tidy_stacks_haplotypes_vcfis now deprecated in favor of usingtidy_genomic_datathat will import haplotypic vcf files.
v.0.4.4
- several updates to make functions faster.
stackr_imputations_moduleno longer imputes globally after imputations by populations. Instead, usecommon.markersor not to test impacts.- bug fix with
change_allelesthat was not working properly inside the imputation module. snp_ldis not a separate module available for users. Check documentation.missing_visualizationnow show the proportion of variance with plot axis text.
For previous news: NEWS.md file
- Until publication stackr will change rapidly (see contributions below for bug reports).
- Updated filters: more efficient, interactive and visualization included: in progress
- Workflow tutorial that links functions and points to specific vignettes to further explore some problems: in progress
- Integration of several functions with STACKS and DArT database.
- Use Shiny and ggvis when subplots or facets becomes available...
- Suggestions ?
This package has been developed in the open, and it wouldn’t be nearly as good without your contributions. There are a number of ways you can help me make this package even better:
- If you don’t understand something, please let me know.
- Your feedback on what is confusing or hard to understand is valuable.
- If you spot a typo, feel free to edit the underlying page and send a pull request.
New to pull request on github ? The process is very easy:
- Click the edit this page on the sidebar.
- Make the changes using github’s in-page editor and save.
- Submit a pull request and include a brief description of your changes.
- “Fixing typos” is perfectly adequate.
The stackr package fits currently at the end of the GBS workflow. Below, a flow chart using STACKS and other software. 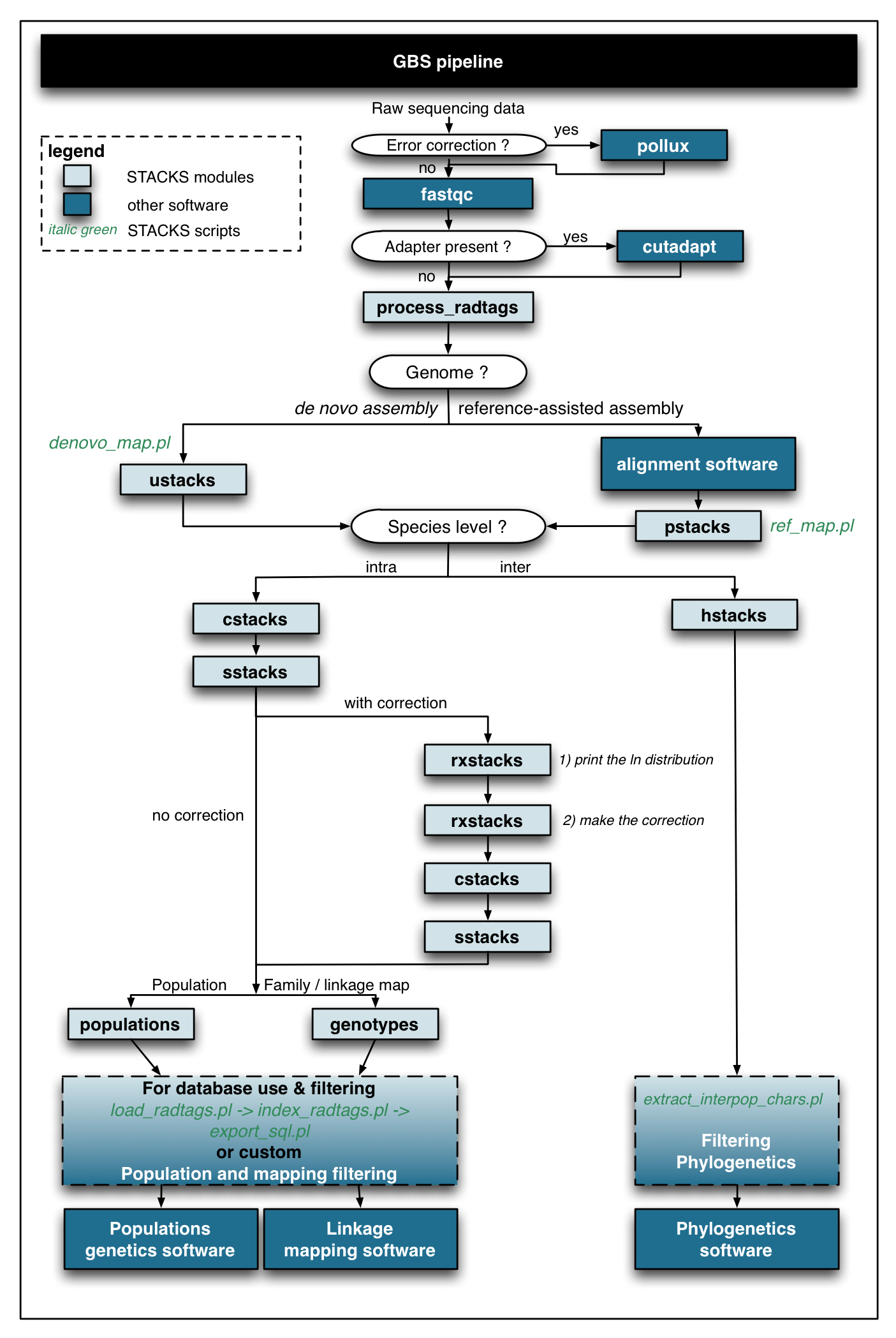
Currently under construction. Come back soon!
Table 1: Quality control and filtering RAD/GBS data
| Parameters | Libraries & Seq.Lanes | Alleles | Genotypes | Individuals | Markers | Sampling sites | Populations | Globally |
|---|---|---|---|---|---|---|---|---|
| Quality | x | x | ||||||
| Assembly and genotyping | x | |||||||
| Outliers | x | x | x | x | ||||
| Missingness | x | x | x | x | x | x | x | x |
| Coverage | x | x | x | |||||
| Genotype Likelihood | x | |||||||
| Prop. Genotyped | x | x | x | x | x | |||
| HET & FIS & HWE | x | x | x | |||||
| MAF | x | x | x | x | ||||
| Missingness | x | x | x | x | x | x | x | x |
Step 1 Quality Ask yourself these questions:
- DNA quality, libraries quality, sequencing lanes quality ?
- Please stop thinking in terms of quantity (e.g. millions of reads returned), and start thinking about actual quality of your new data.
- Use quality metrics inside available software (e.g. fastqc)
Step 2 de novo assembly and genotyping
- This is conducted outside stackr
- Integrated software pipelines include: STACKS, pyRAD, dDocent, AftrRAD. If you want to develop your own pipeline, there are a multitude of approaches, good luck.
Step 3 Outliers
- Remove replicates (I hope you have some).
- Remove de novo assembly artifact, by creating blacklist of genotypes or whitelist of markers:
- individuals with more than 2 alleles (use
summary_haplotypes) - outlier markers with extreme number of SNP/read or haplotype (use
filter_snp_number)
- individuals with more than 2 alleles (use
- Remove potential duplicated samples that went off your radar, try
detect_duplicate_genome. - Highlight outliers individual's heterozygosity that might represent mixed samples with
detect_mixed_individuals. - The metric you're using: a de novo artefact or a reliable signal of biological polymorphism?
- Should the statistic you are interested in be consistent throughout the read ?
- Will you consider haplotype or snp level statistics?
- The consistensies of SNPs statistics among haplotype can be throughly tested by using
snp.ldargument in several stackr functions. - Any other outliers with different individual's or markers metrics (reads/sample, etc) ?
Step 4 Pattern of missingness
- Use
missing_visualizationwith/without your new blacklists (e.g. of genotypes, individuals) and with/without whitelist of markers to examine patterns of missingness in you dataset before more extensive filtering (there is a vignette for this step) - The trick here is to use the
strataargument to find patterns associated with different variables of your study (lanes, chips, sequencers, populations, sample sites, reads/samples, etc). - Do you see a trend between your missing pattern and reads/samples ? Heterozygosity?
- Do you need more sequencing? Do you have to re-run some lanes?
Step 5-6 Coverage and Genotype Likelihood
- Coverage is an individual metric. With most software you'll find allele and genotype coverage info.
- Genotype likelihood is usually a metric based on coverage of the different genotypes found in all of your data.
- Good allele coverage is required for reliable genotypes.
- Reliable genotypes is required for reliable downstream summary statistics.
- Explore filtering options in
filter_coverageandfilter_genotype_likelihood.
Step 7 Prop. Genotyped
- Do you have enough individuals in each sampling sites (
filter_individual) and enough putative populations (filter_population) for each markers ? - Use blacklist of individuals with different thresholds.
- Keep different whitelist of markers.
- Use
common.markersargument inside most of stackr functions to test the impact of vetting loci based on shared markers. - Use imputation methods provided by stackr (inside
tidy_genomic_dataorgenomic_converter, as a separate module:stackr_imputations_module) to assess the impact of lowering or increasing threshold that impact missing data.
Step 8 HET, Fis, HWE
- Overall and/or per populations hwe, heterozygosity and Fis statistics can highlight: de novo assembly problems (oversplitting/undermerging), genotyping problems or biological problems.
- These filters allows to test rapidly if departure from realistic expectations are a problem for downstream analysis ?
- Choose your threshold wisely and test impact on pipeline.
- Use
filter_het,filter_fis,filter_hweand look again at the individual's heterozygosity (detect_mixed_individuals) for outliers.
Step 9 MAF
- Remove artifactual and uninformative markers.
- Use MAF arguments inside several of stackr functions to tailor MAF to your analysis tolerance to minor allelel frequencies.
- There is also a separate filter in stackr:
filter_maf
Step 10 Pattern of missingness, again
- Use
missing_visualizationwith your new blacklists (e.g. of genotypes, individuals) and with your whitelist of markers to examine patterns of missingness in your dataset after filtering (there is a vignette for this step) - The trick here is to use the
strataargument to find patterns associated with different variables of your study (lanes, chips, sequencers, populations, sample sites, reads/samples, etc). - Do you see a trend between your missing pattern and reads/samples ? Heterozygosity?

