Deep Geometric Representations for Modeling Effects of Mutations on Protein-Protein Binding Affinity
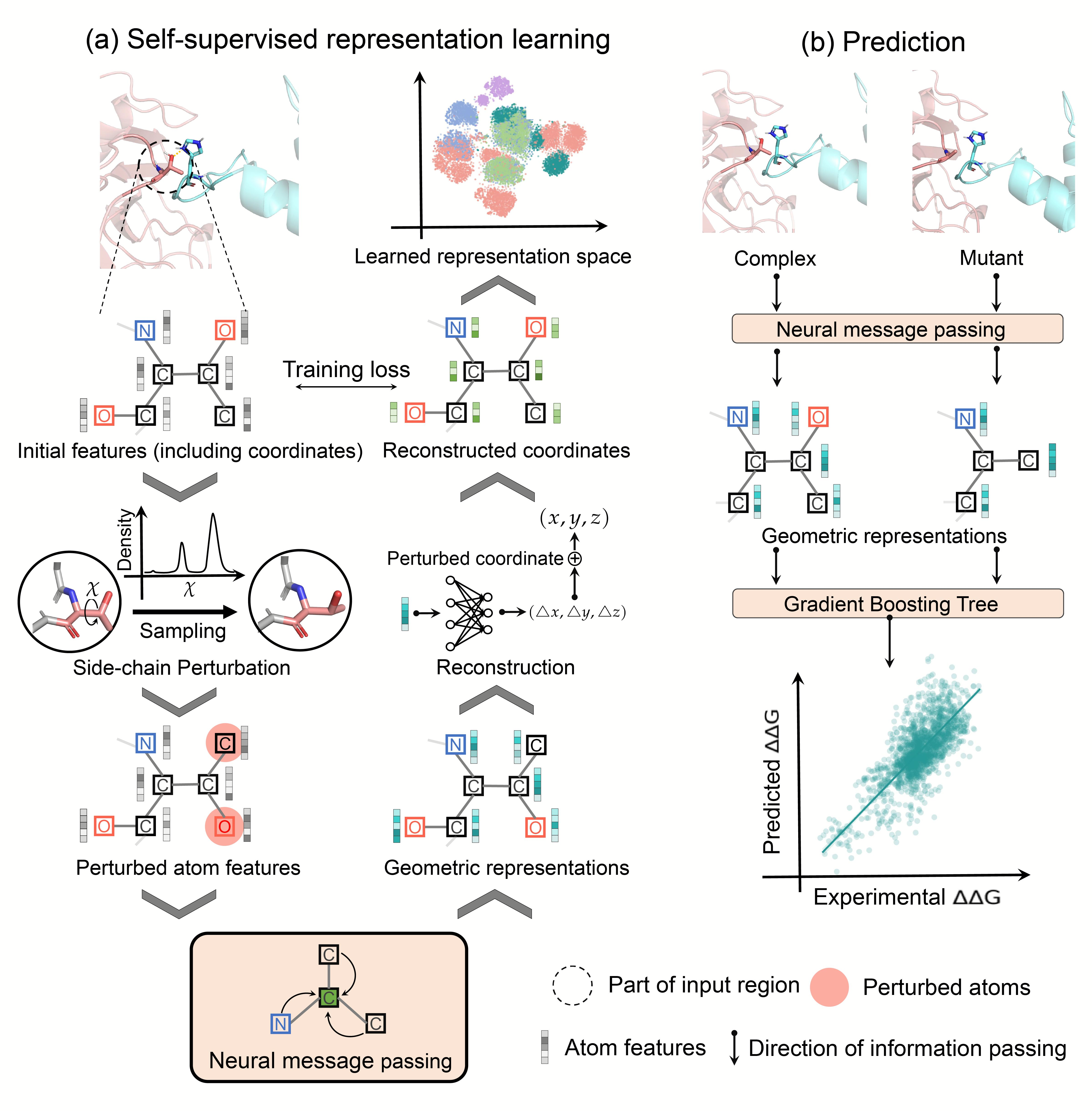
GeoPPI is a deep learning based framework that uses deep geometric representations of protein complexes to model the effects of mutations on the binding affinity. To achieve both the powerful expressive capacity for geometric structures and the robustness of prediction, GeoPPI sequentially employs two components, namely a geometric encoder (excelling in extracting graphical features) and a gradient-boosting tree (GBT, excelling in avoiding overfitting). The geometric encoder is a graph neural network that performs neural message passing on the neighboring atoms for updating representations of the center atom. It is trained via a novel self-supervised learning scheme to produce deep geometric representations for protein structures. Based on these learned representations of both a complex and its mutant, the GBT learns from the mutation data to predict the corresponding binding affinity change.
Thanks to the above design, GeoPPI enjoys accurate predictive power, strong generalizability, and high inference speed for the estimation of the mutation impact.
This source code is tested with Python 3.8 onUbuntu 20.04. Users need to accomplish the following three steps to complete the installation.
git clone https://github.com/Liuxg16/GeoPPI.git
cd GeoPPIBuilding the required dependencies requires runing the script:
source install.sh [flag]If your system has installed Anaconda software, please set [flag] to 1, otherwise set [flag] to 0.
The above script will complete the following two things: 1) building a virtual enviroment named "ppi"; 2) installing required dependencies. If you meet any difficulty during this installation, please refer to the full documentation (i.e., GeoPPI documentation.pdf) for more details.
The FoldX Suite is available through academic and commercial licenses. Please apply for a license and download FoldX v4.0 binary file from: http://foldxsuite.crg.eu/
Once you download the FoldX file, please unzip the file and put the FoldX binary file in this main directory (i.e., GeoPPI/foldx). For example, suppose the file name is "foldxLinux64.tar.gz", run the following commands (ubuntu environment):
cp foldxLinux64.tar.gz ./
tar -zxvf ./foldxLinux64.tar.gz
chmod a+x ./foldxCongratulations! The environment is ready to run GeoPPI.
Users can use GeoPPI to compute the binding affinity changes given the complex and the mutation information.
Before using GeoPPI, please activate the environment first.
conda activate ppiThen, you can use the following command to obtain the results:
python predict.py [pdb file] [Mutation] [partnerA_partnerB]where [pdb file] is the complex structure of interest, [Mutation] denotes the mutation information and [partnerA_partnerB] describes the two interaction partners in the protein complex.
Format of [Mutation]: The mutation information includes WT residue, chain, residue index and mutant residue. such as “TI38F”, which stands for mutating the 38th acid amino at the I chain (i.e., phenylalanine) to threonine.
Format of [partnerA_partnerB]: [partnerA_partnerB] are the chains of the two parts of the binding. For example, if the H chain and the A chain of the complex belong to different proteins and interact with each other in the complex, [partnerA_partnerB] is “A_H”. Similarly, “HL_WV” stands for the H and L chains interact with the W and V chains.
Program output: After several seconds of computing, the GeoPPI program will return the impact of the input mutation, i.e.,
Thererfore, the positive value stands for the higher binding affinity between two proteins, i.e., the stabilizing mutation.
For example, when we execute the command:
python predict.py data/testExamples/1PPF.pdb TI17F E_IThe program output is similar to the following:
========================================Results============================================
The predicted binding affinity change (wildtype-mutant) is -1.76 kcal/mol (destabilizing mutation).In the GeoPPI/data directory, there are several example complexes for users to test GeoPPI. Here, we also provide some example commands as follows.
python predict.py data/testExamples/1PPF.pdb TI17R E_I
python predict.py data/testExamples/1CZ8.pdb KW84A WV_HL
python predict.py data/testExamples/1CSE.pdb LI38I E_I
python predict.py data/testExamples/3SGB.pdb KI7L E_I
python predict.py data/testExamples/3BT1.pdb PU149A U_AUsers can also use their own structures to analyze the mutation effects by putting the PDB files into the directory data/testExamples/ and executing the above command again:
python predict.py [pdb file] [Mutation] [partnerA_partnerB]If you encounter any problems during the setup of environment or the execution of GeoPPI, do not hesitate to contact liuxg16@mails.tsinghua.edu.cn or create an issue under the repository: https://github.com/Liuxg16/GeoPPI.
Cheers!