UDiPH is a package that creates a new metric space where the points are uniformly sampled with respect to a global metric. It is oriented for creating Vietoris-Rips filtrations that are independent of the metric of the initial space. This work is very influced on UMAP.
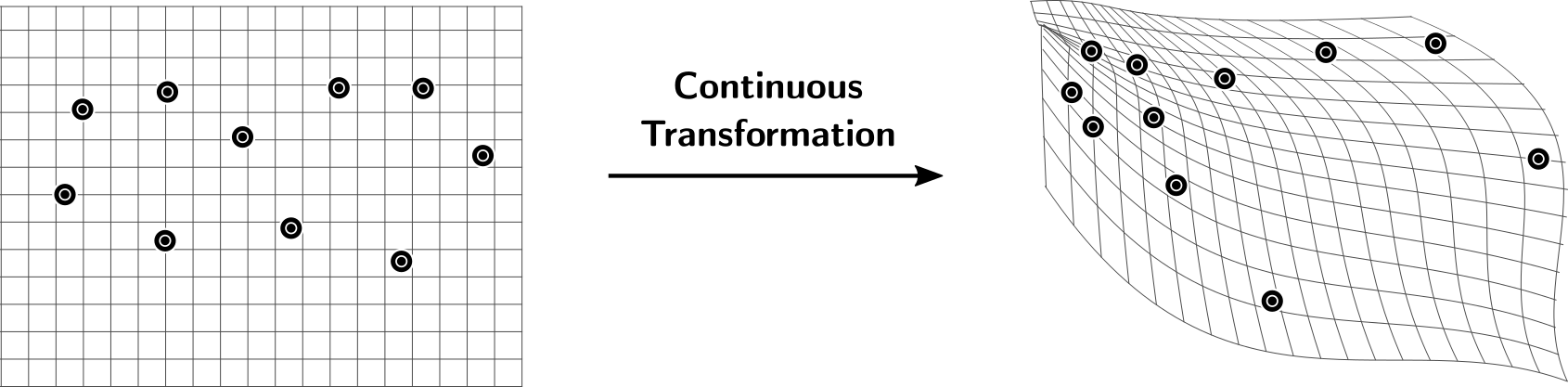
UDiPH excells when comparing homology of spaces with different metrics, for example consider a continuous deformation acting on a set of points:
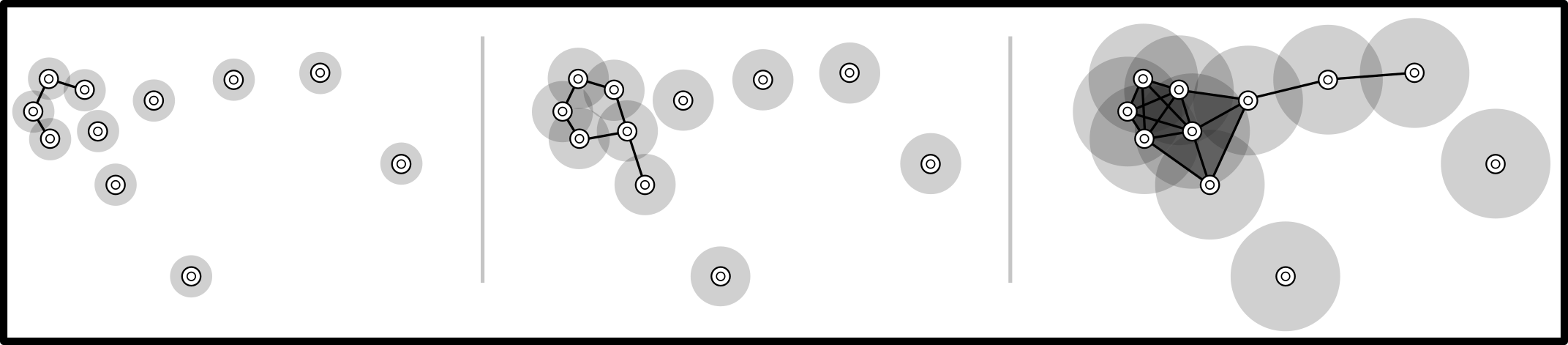
Standard Vietoris-Rips filtration:
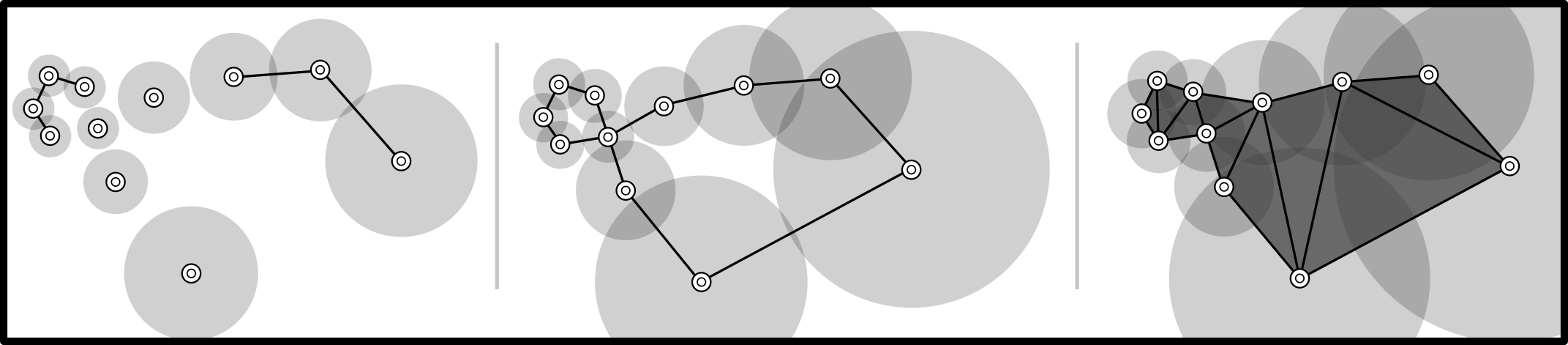
Vietoris-Rips filtration using metric space created by UDiPH is based on local density: Since we are in the presence of a continuous deformation it is expected that the homology remains invariant. However due to the difference in metric in the image space, the standard Vietoris-Rips filtration does not accuratly capture this invariance. Notice how a filtration using UDiPH manages to capture both homology generators.Dependencies:
- Numpy
- Scipy
- Sklearn
UDIPH is available on PyPI,
pip install udiphimport udiph
M = udiph.UDIPH(X=X, n_neighbors=15, distance_matrix=False, return_complex=False)Parameters:
X: numpy array of shape=(samples,features) or shape=(samples,samples) containing the data. If data is a pairwise distance matrix thendistance_matrixmust be set to True.n_neighbors: number of nearest neighbors considered when creating the proximity graph. Too many and topological features are dissolved, too few are artifacts are created. Fairly robust.distance_matrix: Boolean value indicating if input data is or not a distance_matrixreturn_complex: Boolean value indicating whether to return the weighted 1-d simplicial complex instead of a pairwise distance matrix.
Returns:
M: numpy array (samples, samples) pairwise distance matrix with respect to new Riemannian metric.A: (optional) numpy array (samples, samples) adjacency matrix of the weighted 1-d simplicial complex.
The basic assumption on UDiPh relies on the idea that data comes uniformly sampled from an unknown Riemmannian manifold. As a consequence, "bigger" holes in the governing manifold are respresented by having a higher number of points sampled from their boundary, and "small" holes will have less points sampled.
If you use UDiPH in your work or parts of the algorithm please consider citing:
Paper coming soon