This repo has been moved to the CogStack organisation. No further updates will be made to this repo
This repository contains all tools relevant to interacting with an NHS deployment of CogStack.
It contains easy to follow templates and instructions to interact and search CogStack.
NOTE These scripts have been moved to the main repo in the CogStack Org
Tasks left TODO:
- CogStack Search
- CogStack Watcher Jobs
- MedCAT creating a model
- MedCAT unsupervised training a model
- MedCAT supervised training a model
- MedCAT annotating free text
Users can follow these steps to quickly setup and deploy this repository on their machine.
Any code to enter in these instructions will be represented as code to enter.
Please replace anything within <Enter information here> with your own specific details.
-
Enter the directory where you would like to store these files.
cd path/to/where/you/want/this/repository -
Clone the online repository:
git clone https://github.com/antsh3k/working_with_cogstack.gitFurther instructions and self-help with git and git clone. Please visit this link.
If you choose to use github desktop rather than the terminal please refer to the official github desktop guides.
-
Optional: To update to the latest release of this repository:
git pull
(Requires Python 3.7+)
Windows
- Create a new virtual env:
python3 -m venv venv - Load the virtual environment:
.\venv\Scripts\activate - Install relevant packages and libraries:
pip install -r requirements.txt
Linux/MAC OS
- Create a new virtual env:
python3 -m venv venv - Load the virtual environment:
source venv/bin/activate - Install relevant packages and libraries:
pip install -r requirements.txt
Optional: If no jupyter instance is installed.
- In the main folder of this repository. Activate your virtual environment, using the (Step 2) command from your respective OS.
- Start jupyter notebook:
jupyter notebook
In the main folder of this repository you must populate the credentials.py file with your own CogStack hostnames, username and passwords.
If you have any questions or issues obtaining these details please contact your local CogStack administrator.
The credentials.py contents:
hosts = [] # This is a list of your cogstack elasticsearch instances.
# These are your login details (either via http_auth or API)
username = ''
password = ''
api_username = ''
api_password = ''
This directory contains the basics search templates.
For further information on CogStack please visit their github or wiki page.
This directory contains the basics watcher job templates.
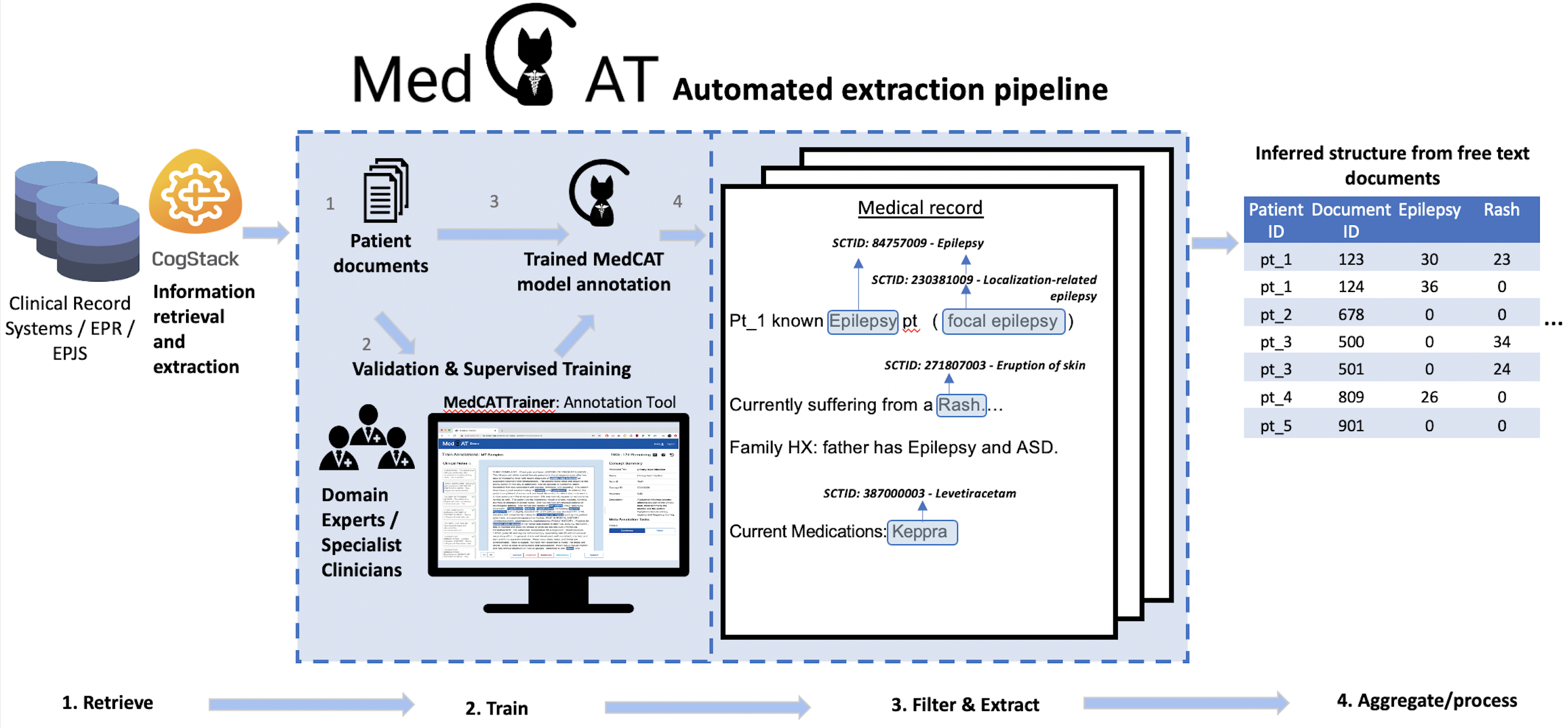
An overview of this process is shown below.
Further information about MedCAT can be found from their github or via their official documentation here.
General MedCAT tutorials can be found here.
A demo application is available at MedCAT. This was trained on MIMIC-III to annotate SNOMED-CT concepts. Note: No supervised training has been provided to this model and therefore should only be used for demonstration purposes only.
@ARTICLE{Kraljevic2021-ln,
title="Multi-domain clinical natural language processing with {MedCAT}: The Medical Concept Annotation Toolkit",
author="Kraljevic, Zeljko and Searle, Thomas and Shek, Anthony and Roguski, Lukasz and Noor, Kawsar and Bean, Daniel and Mascio, Aurelie and Zhu, Leilei and Folarin, Amos A and Roberts, Angus and Bendayan, Rebecca and Richardson, Mark P and Stewart, Robert and Shah, Anoop D and Wong, Wai Keong and Ibrahim, Zina and Teo, James T and Dobson, Richard J B",
journal="Artif. Intell. Med.",
volume=117,
pages="102083",
month=jul,
year=2021,
issn="0933-3657",
doi="10.1016/j.artmed.2021.102083"
}