This repo contains codes needed for NMF based on MU and HALS. It implements algorithms from the paper https://arxiv.org/abs/1107.5194
This codes require numpy, cv2 and matplotlib for NMF and plots part.
Deep learning models require tensorflow, tritonclient, protobuf. These packages are not needed for run.py.
Two NMF algorithms are implemented in nmf.py, if alpha=0, they reduce to non-accelerated versions.
# Accelerated Multiplicative Update
accelerated_MU(X, L, alpha, epsillon, max_iter, update_func="gillis", save_every_iter=-1, save_with_index=False, save_cb=None, weight_path="", restart=False)# Accelerated Hierarchical ALS
accelerated_HALS(X, L, alpha, epsillon, max_iter, update_func="gillis", save_every_iter=-1, save_with_index=False, save_cb=None, weight_path="", restart=False)Main codes contained in run.py.
python run.py --help
usage: run.py [-h] [--datadir DATADIR] [--algorithm ALGORITHM] [--accelerated]
[--update-func UPDATE_FUNC] [--L L] [--alpha ALPHA] [--eps EPS]
[--max-iter MAX_ITER] [--outputdir OUTPUTDIR]
[--output-recon-images] [--use-sample] [--restart]
[--save-every SAVE_EVERY] [--save-with-index] [--seed SEED]
Accelerated MU/HALS
optional arguments:
-h, --help show this help message and exit
--datadir DATADIR path to directory containing standardized data
(default: None)
--algorithm ALGORITHM
NMF algorithm to use, MU or HALS (default: None)
--accelerated whether or not to use the accelerated version
True/False (default: False)
--update-func UPDATE_FUNC
which update function to use for hals, `paper` or
`gillis` (default: gillis)
--L L low rank dimension (default: 30)
--alpha ALPHA control parameter for number of inner loop iterations,
ignored if accelerated=False (default: 2)
--eps EPS control parameter for stopping condition, ignored if
accelerated=False (default: 0.1)
--max-iter MAX_ITER number of iterations to update W and H (default: 2)
--outputdir OUTPUTDIR
where to export the output (default: None)
--output-recon-images
if true, will generate reconstructed images (default:
False)
--use-sample if true, make use of the list in meta.py to output
report (default: False)
--restart if true, restart from files available in outputdir
(default: False)
--save-every SAVE_EVERY
save weight every # iterations (default: 0)
--save-with-index save weight every # iterations, without replacement
(default: False)
--seed SEED random seed (default: 42)Commands to run can be found in script_all.sh, e.g. to perform NMF with L=30 using accelerated HALS for 100 iterations:
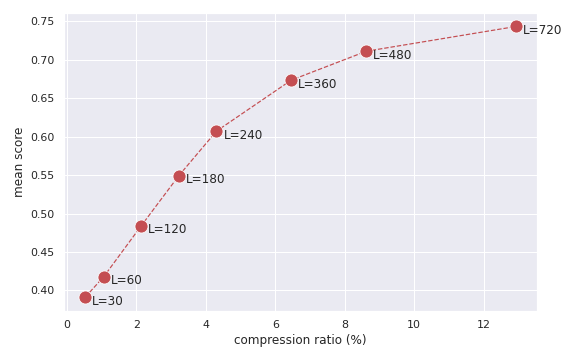
python run.py --datadir /mnt/i/dataset-processed --algorithm hals --accelerated --update-func gillis --L 60 --alpha 0.5 --eps 0.1 --max-iter 100 --outputdir ./HALS_Accel_L60Other Python files help generating some figures, such as compression ratio