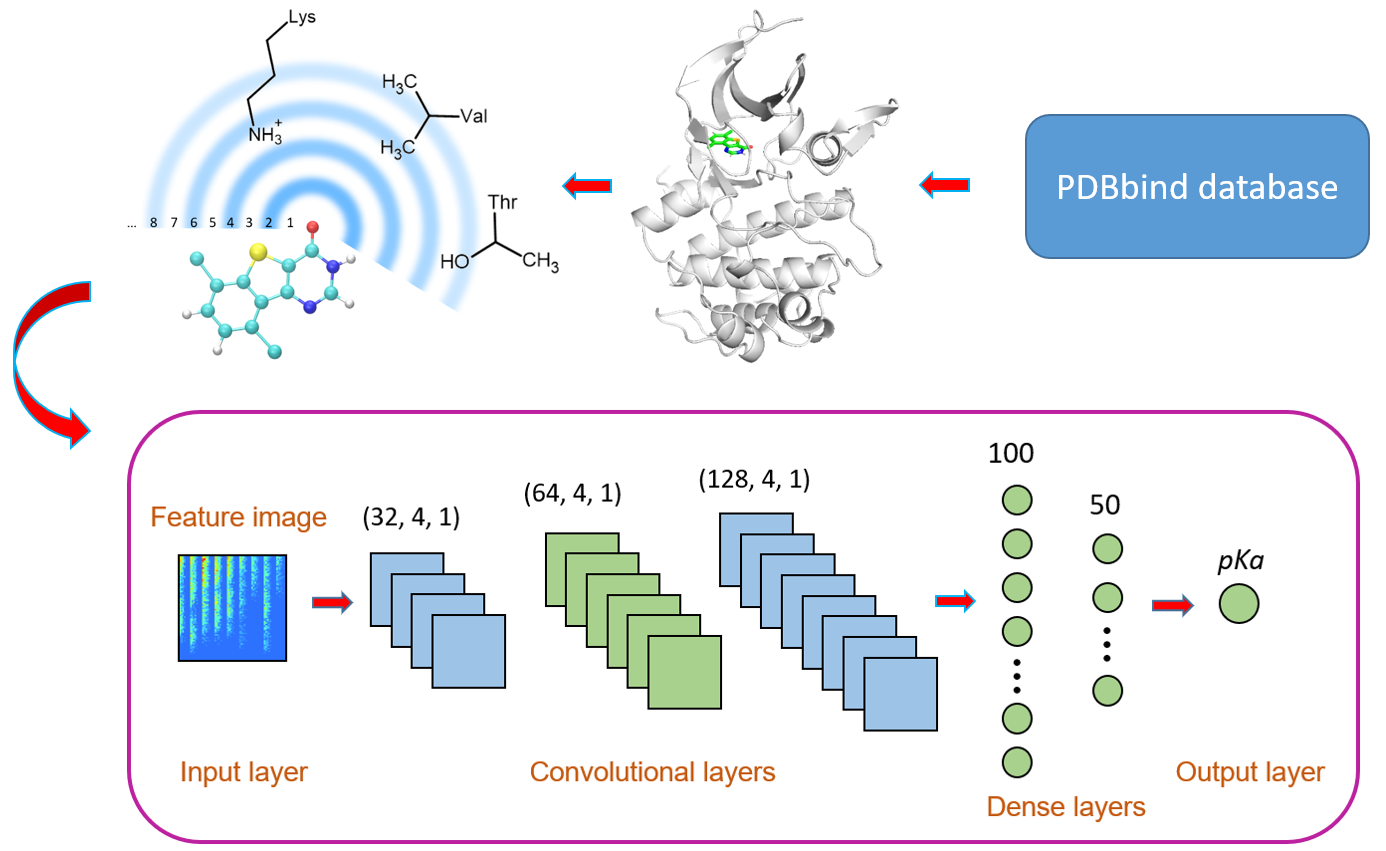
OnionNet-2
OnionNet-2 is constructed based on convolutional neural network (CNN) to predict the protein-ligand binding affinity. One of the greatest advantages of OnionNet-2 is that it can achieve higher accuracy at a lower computational cost. When taking CASF-2016 and CASF-2013 as benchmark, OnionNet-2 shows strong scoring power.
Contact
Prof. Weifeng Li, Shandong University, lwf@sdu.edu.cn
Zechen Wang, Shandong University, zechenwang@mail.sdu.edu.cnCitation
Installation
First, create a conda environment and install some necessary packages for running OnionNet-2.
conda create -n OnionNet-2 python=3.8
conda activate OnionNet-2
conda install numpy
conda install pandas
conda install scikit-learn=0.22.1
conda install -c openbabel openbabel
conda install -c conda-forge mdtraj
Or, you can also use pip to install above packages. For example,
pip install tensorflow-gpu==2.3
Usage
1. Prepare the PDB file containing the 3D structure of protein-ligand complexes.
In the samples/prepare_complexes directory, we provide two scripts to conveniently prepare the PDB file containing 3D structure of the protein-ligand complex.
# Specify the relative path or absolute path of the working directory correctly. For example,
bash prepare_PDB_file.sh ..
2. Generate the residue-atom contact features.
python generate_features.py -h
python generate_features.py -shells N -inp inputs_complexes.dat -out output_featurs.csv
The input file (inputs_complexes.dat) contains the path of the protein-ligand complexes pdb files, for example
1a30/1a30_complex.pdb
1bcu/1bcu_complex.pdb
3. Train the convolutional neural network.
# The features and the real pKa are concated in a common file as the input of the training process. We can execute this process with a script.
python concat_features_pKa.py -inp_features output_features.csv -inp_true all_complexes_pKa.csv -out output_features_pKa.csv
# The training process will output 3 files, the default are "logfile", "bestmodel.h5" and "train_scaler.scaler", of which the latter two will be the input of the prediction process.
python train.py -h
python train.py -train_file train_features_pKa.csv -valid_file valid_features_pKa.csv -shape 84 124 1
4. Predict the protein-ligand binding affinity.
python predict_pKa.py -h
python predict_pKa.py -scaler train_scaler.scaler -model bestmodel.h5 -inp input_features.csv -out predicted_pKa.csv