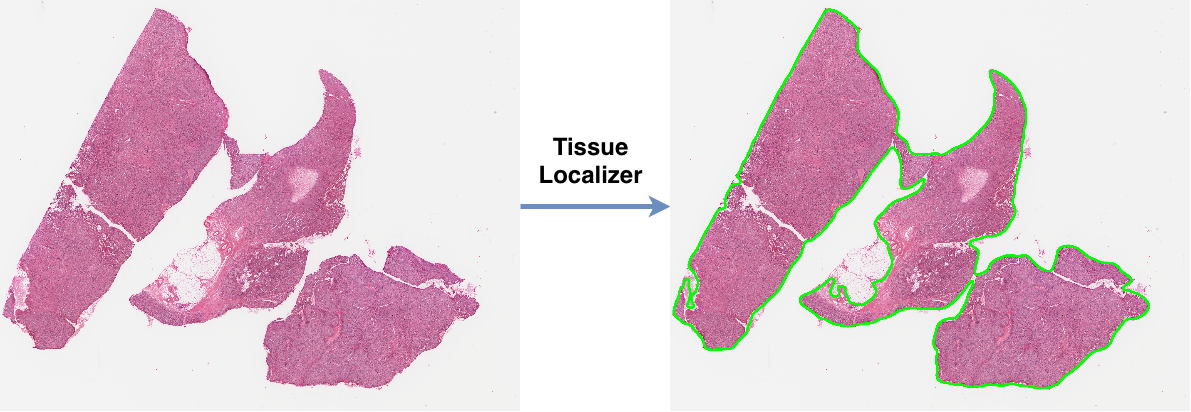
- Install OpenSlide.
$ apt-get install openslide-tools
- Install tissueloc.
$ pip install tissueloc
def locate_tissue_cnts(slide_path,
max_img_size=2048,
smooth_sigma=13,
thresh_val = 0.80,
min_tissue_size=10000):
""" Locate tissue contours of whole slide image
Parameters
----------
slide_path : valid slide path
The slide to locate the tissue.
max_img_size: int
Max height and width for the size of slide with selected level.
smooth_sigma: int
Gaussian smoothing sigma.
thresh_val: float
Thresholding value.
min_tissue_size: int
Minimum tissue area.
Returns
-------
cnts: list
List of all contours coordinates of tissues.
d_factor: int
Downsampling factor of selected level compared to level 0
"""
Testing slide can be downloaded from Figshare.
import tissueloc as tl
slide_path = "../data/SoftTissue/TCGA-B9EB312E82F6.svs"
# locate tissue contours with default parameters
cnts, d_factor = locate_tissue_cnts(slide_path,
max_img_size=2048,
smooth_sigma=13,
thresh_val=0.80,
min_tissue_size=10000)
import tissueloc as tl
# Step 1: Select the proper level
slide_path = "../data/SoftTissue/TCGA-B9EB312E82F6.svs"
max_img_size = 2048
s_level, d_factor = tl.select_slide_level(slide_path, max_img_size)
# Step 2: Load Slide image with selected level
slide_img = tl.load_slide_img(slide_path, s_level)
# Step 3: Convert color image to gray
gray_img = tl.rgb2gray(slide_img)
# Step 4: Smooth and Binarize
thresh_val = 0.8
smooth_sigma = 13
bw_img = tl.thresh_slide(gray_img, thresh_val, sigma=smooth_sigma)
# Step 5: Fill tissue holes
bw_fill = tl.fill_tissue_holes(bw_img)
# Step 6: Remove small tissues
min_tissue_size = 10000
bw_remove = tl.remove_small_tissue(bw_fill, min_tissue_size)
# Step 7: Locate tissue regions
cnts = tl.find_tissue_cnts(bw_remove)
Hosted in https://tissueloc.readthedocs.io, powered by readthedocs and Sphinx.
tissueloc is free software made available under the MIT License. For details see the LICENSE file.
See the AUTHORS.md file for a complete list of contributors to the project.
Issues and Pull Request are very welcome.