Machine learning methods utilizing various structure-based and energy-based descriptors characterizing protein-ligand interactions to predict activity for novel compounds against a broad range of human kinases.
Predicting models are trained on the dataset of 104 human kinases with available PDB structures and with available experimental activity data against 1202 small-molecule compounds from PubChem BioAssay dataset ‘Navigating the Kinome’ (https://pubchem.ncbi.nlm.nih.gov/bioassay/493040#section=Top).
The project contains Jupyter notebook scripts for models training, separate scripts for models evaluating, saved files with pre-trained models, bash-script for the pipeline to prepare descriptors for machine learning.
Kinase-ligand complexes in our pipeline are obtained with SMINA docking software and docking scores are used as part of the descriptors set.
Descriptors preparation pipeline utilizes a number of tools and software, including
Most of these tools are freely available except for ICM-Pro, also it was used only on the steps 1 and 3 for a few operations and could be replaced by any other suitable programs. We used ICM in our pipeline particularly to save protein-ligand complex structures in PDB files after docking for further analysis and to calculate protein-ligand interaction surface (ICM_area), number of protein-ligand hydrogen bonds (ICM_hbonds), number of atoms in ligands (nof_Atoms), number of rotational bonds in ligands (nof_RotB).
Descriptors preparation pipeline can be run as bash run_pipeline.sh, please correct pathways to software in a header of the script.
Pipeline contains the following steps:
- Calculate nof_Atoms and nof_RotB for ligands with ICM-Pro.
- Calculate solvation energy for free ligands.
- Prepare PQR files for ligands to run r6_born utility with Amber tools and ACPYPE script.
- Run GB_NSR6 utility to calculate solvation energy of free ligands.
- Run docking with SMINA and process docking results with ICM-Pro.
- Re-score protein-ligand complexes with X-score.
- Analyze protein-ligand contacts with PLIP (pyMol based tool).
- Join tables into summary descriptors table 'test_compl_descr.tab' with R.
Kinases PDB files that should be used for docking can be found in recept.tar.gz. Receptors list is available in file recept.tab. Number of descriptors associated with receptors are already pre-calculated and available in tarSim_allPairs.tab
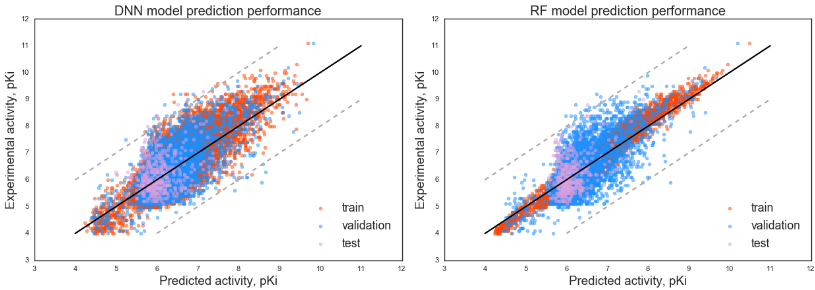
The main script for activity prediction models' training can be found in Jupyter notebook file train_main_model.ipynb, the script contains descriptors' processing and construction and training of the two activity predicting models: DNN model, built with Keras, and Random Forest model built with Scikit-learn library.