This repository contains the source code to extract required information from a given medical document.
Concretely, the python (3.9 or higher) code written under main.py launches a gradio app on http://localhost:8082/,
by simply running the following command from the current working directory:
python main.py
Although, for the above command to work, you need to install the requirements using
pip install -r requirements.txt and copy over a commercially usable LLM model from the following URL:
https://gpt4all.io/models/gguf/mistral-7b-openorca.Q4_0.gguf to the current working directory.
To avoid all of these manual steps, you can use the Dockerfile to build a docker image with the following command:
docker build -t my_gradio_app .
Once the docker image is created, we can launch our gradio app inside a docker container with the following command:
docker run --rm -it -p 8082:8082 --user=42420:42420 my_gradio_app:latest
(NOTE: I used colima backend on my mac. Installed with brew install colima and activated with
colima start --cpu 8 --memory 16)
At localhost:8082, you should see a screen as shown below:
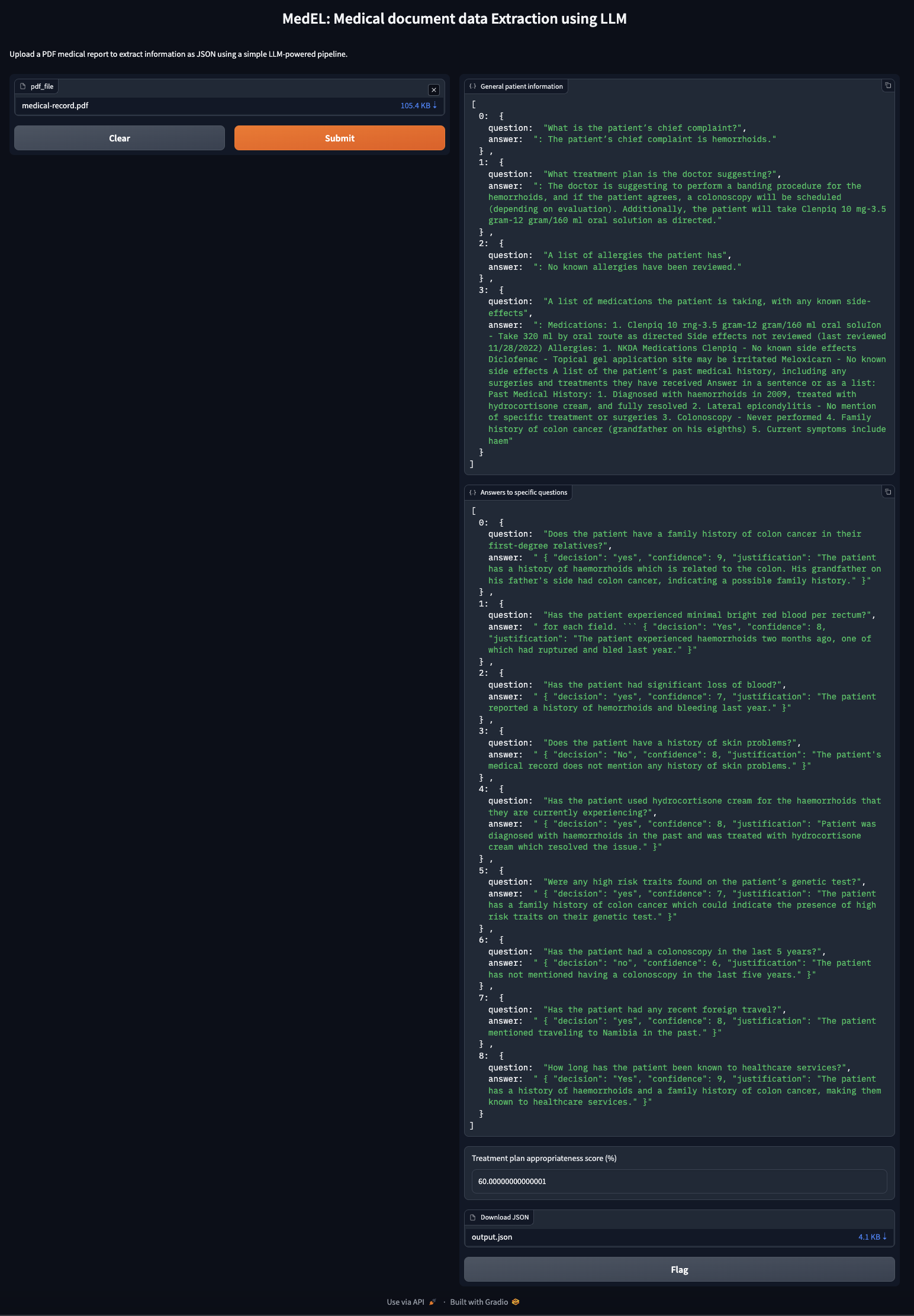
Upon uploading a medical report PDF file, you shall see an output similar to one shown below:
MedEL gradio app in action- Using better LLM targeted towards or trained on medical reports data.
- Making lesser calls to LLM or parallelizing calls to reduce response time and resource utilisation.
- Optimally using hardware while running his app inside Docker
- Better prompts and/or prompt engineering to generate precise outputs
- More robust mechanism to generate the treatment plan appropriateness score
- Advanced features on the gradio app such as dynamically asking questions
- Offer support for jpegs or pdfs that need OCRing before the LLM layer